This photo taken on Oct. 21, 2025 shows the comet C/2025 A6 (Lemmon) in the sky over Tangyuan County, northeast China’s Heilongjiang Province. (Photo by Zhu Zongqiang/Xinhua)

This photo taken on Oct. 20, 2025 shows the comet C/2025 A6 (Lemmon) and…

This photo taken on Oct. 21, 2025 shows the comet C/2025 A6 (Lemmon) in the sky over Tangyuan County, northeast China’s Heilongjiang Province. (Photo by Zhu Zongqiang/Xinhua)

This photo taken on Oct. 20, 2025 shows the comet C/2025 A6 (Lemmon) and…
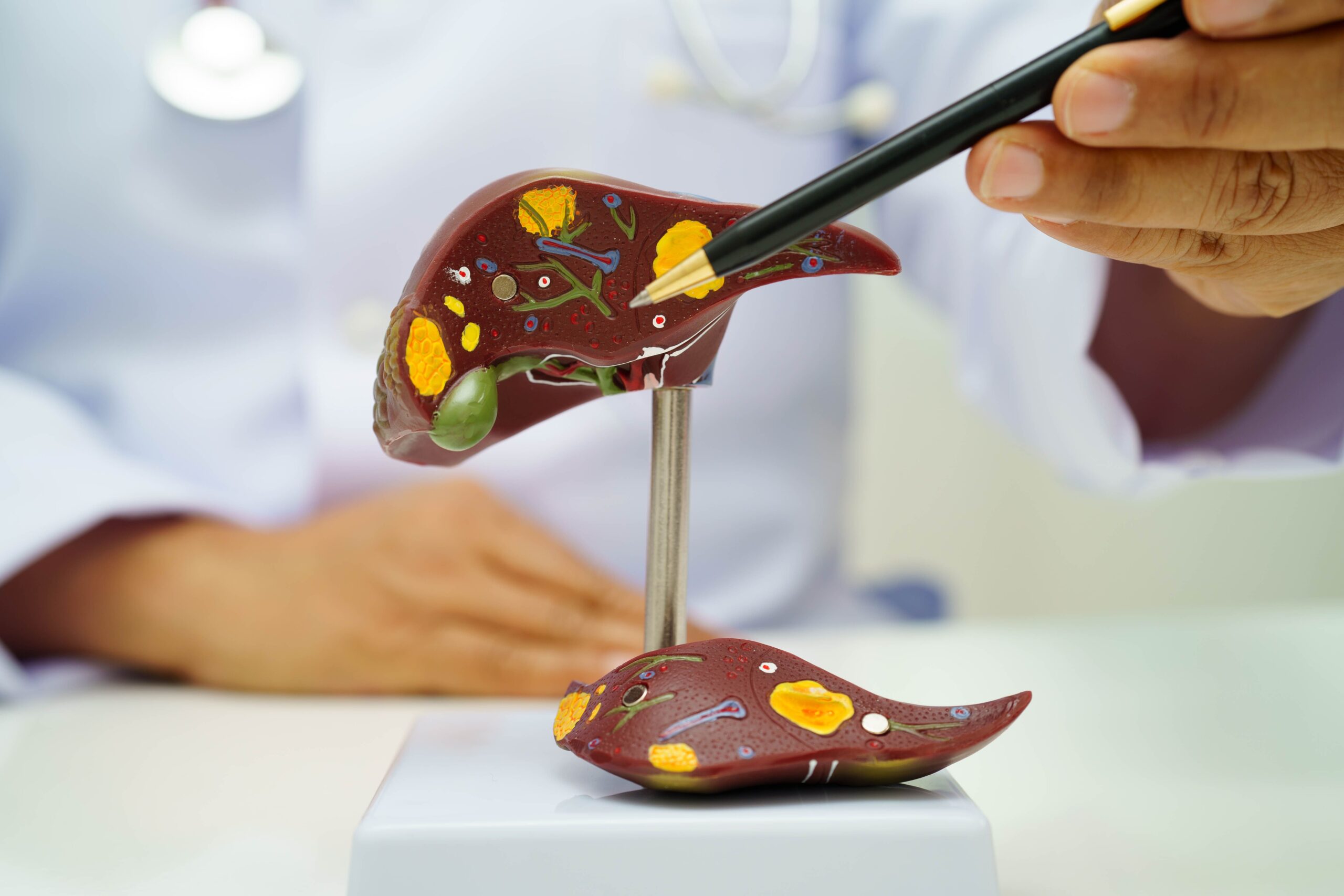
The researchers used National Inpatient Samples from 2016 to 2020, identifying the hospitalizations involving diagnoses of MASLD, the disease formerly known as nonalcoholic fatty liver disease, and MASH, then compared these groups to patients…
The same Nobel-winning mRNA technology that helped curb the Covid-19 pandemic may be poised to transform cancer care.
Patients with advanced lung or skin cancer who received a Covid-19 mRNA vaccine within 100 days of starting immunotherapy drugs…

CNBC correspondent Ernestine Siu attended an AI hackathon.
Courtesy of Ernestine Siu
I like to spend most of my weekends taking workout classes with friends, exploring new restaurants or vegging out on the couch with a movie on. The past weekend, however, was unique. Instead of the usual routine, I chose to immerse myself in a 24-hour ‘vibe coding’ hackathon.
I attended one of Singapore’s biggest in-person hackathons ever, which was sponsored and supported by AI heavyweights from around the world including OpenAI, Cursor, Anthropic, Google DeepMind and more.
The event took place on a university campus from about 9 a.m. on Saturday to Sunday noon. Over 400 people participated, ranging from highly experienced engineers to first-timers. The goal was simple: build something from scratch with the help of artificial intelligence.
“Use your imagination… build something unhinged. Build something wacky,” said Agrim Singh, one of the event’s organizers and the co-founder of Niyam AI.
Several teams really took that advice to heart.
Some standout projects included F**Yu.AI, an AI-enabled productivity app that “bullies you into greatness,” by calling users on their cell phones to yell at them to complete tasks, and RizzedIn, a dating site that helps connect “career-minded individuals.”
Some members of the winning teams with CNBC correspondent, Ernestine Siu (right).
Courtesy of Nicholas Cheng
By the end of the weekend, about 150 projects were submitted. The winner – an AI-powered whiteboarding tool that allows users to draw using just their hand movements tracked through a webcam — took home prizes worth over $50,000 Singapore dollars (about $39,000).
The second and third place winners created a human-versus-AI speed game and a “Netflix for corporate training” tool that makes compliance training videos more engaging for employees.
As someone without a technical background, I was nervous entering the event but excited to meet the hackathon community. I knew I had to optimize my chances of building something successfully by teaming up with people who were much more technical than me.
Through Discord, I found my team for the hackathon: Gabrielle Ong who has a product development background, Aung Maung who has a deep tech background, Ninna Cao who has a product design background and Jay Chen who has a software engineering background. What did I offer? Ideas, my storytelling abilities and vibes.
Participants filled a lecture hall at the Singapore University of Technology and Design campus during the hackathon.
Courtesy of Nicholas Cheng
We bonded over a shared desire to preserve the legacies of our grandparents. So, we built Heirloom — a digital time capsule which can be used to capture and store family stories and recipes, so that they can be passed down for generations to come.
We spent all of Saturday working on transforming our idea into a product. Some of us stayed up the entire night working on the project — while others (me) unfortunately didn’t have the stamina to do so.
We were trying to do a bit of a count … And we found people in like random lecture rooms. It was just the funniest [thing] … It was like catching wild Pokemon.
Sherry Jiang
Co-founder, Peek
When I got back to the campus on Sunday morning, I saw people sleeping on benches, on the floor and just about everywhere, while others were still making last minute adjustments before submitting their projects for judging.
An estimated 70 people stayed overnight to work on their project, said Sherry Jiang, one of the event organizers and the co-founder of fintech app Peek.
“We were trying to do a bit of a count … And we found people in like random lecture rooms. It was just the funniest [thing] … It was like catching wild Pokemon,” said Jiang.
Richard Lee, who built a gamified habits-training app Orbie alongside his teammate Amanda Lau Shernin, slept for only about half an hour on the floor of a lecture hall. He had prior developer experience and knows coding languages like Python and SQL.
Participants of the hackathon ranged from highly experienced engineers to newbies.
Courtesy of Nicholas Cheng
When asked why he chose to join the hackathon, he said: “I took it as a personal challenge to see [what] could actually be done within 24 hours … [and to] see how far vibe coding has advanced,” said Lee. He also thought it would be a great place to get inspiration.
“It’s like a gathering of builders … who don’t just learn, but do,” Lee added. “It’s almost like training for a startup, right? Effectively, you just have to focus and get something done.”
By the end of the weekend, Lee says that although the hackathon was just 24 hours, he felt like he had “significantly upgraded” his skills.
The hackathon’s organizers said their goal was to reinvigorate the builder community in Singapore. There’s a feeling among many people in the space that the hackathon scene has faded from its heyday, co-organizer Jiang said.
Fellow organizer Singh agreed, writing on a LinkedIn post that “the Singapore hackathon scene lost its soul.”
Singh, who has been attending hackathons since 2013, observed that such events used to center hackers and builders making “something that worked”, instead of “panels or sponsorship decks or photo ops.”
“Now? Most AI events here feel hollow. Panels by people who’ve never touched the tech. ‘Thought leadership’ with no practical weight. People pretending to build, or worse, extracting revenue from the hype without caring about the ecosystem,” he wrote.
With the advancement of AI, the startup landscape and software engineering industry has changed massively — it’s now easier for people with non-technical backgrounds to build tech products themselves.
Out of all of the participants, about half of them were completely new to hackathons, Jiang said.
Jiang also pointed out that some participants who had learned how to vibe code just a couple weeks ago had placed “pretty high” on the event’s rankings, beating experienced engineers.
“This is a bit of a hypothesis I have … I feel like people who [have a good] product sense and good taste and know how to position their products are starting to do really well at these hackathons because engineering is a lot easier now,” she said. “We’ve lowered the barrier, but raised the bar.”
The time to build is far shorter than [before]. I think it’s really much easier for developers or technical people, or even non-technical people to build a prototype, and effectively get to market.
Richard Lee
Hackathon participant
“The time to build is far shorter than [before]. I think it’s really much easier for developers or technical people, or even non-technical people to build a prototype, and effectively get to market,” said hackathon participant Lee.
Additionally, Lee and Jiang agree that both startup teams and corporate developer teams will likely become smaller now that these AI-assisted coding tools are on the market.
“It’s improving so fast that if [you] are not using [these tools] it day to day … I think [you’ll] be at a big risk of being eliminated.”
Richard Lee
Hackathon participant
With that said, it’s still very helpful to have some background on software engineering, as this will carry you to the finish line. People will still need to know how to evaluate code in terms of its fundamental logic and understand how to address issues in code, hackathon participant Lee said.
Ultimately, AI models are improving so quickly that if engineers aren’t upskilling consistently on the tools and staying up to date, they are at risk of becoming redundant, he added.
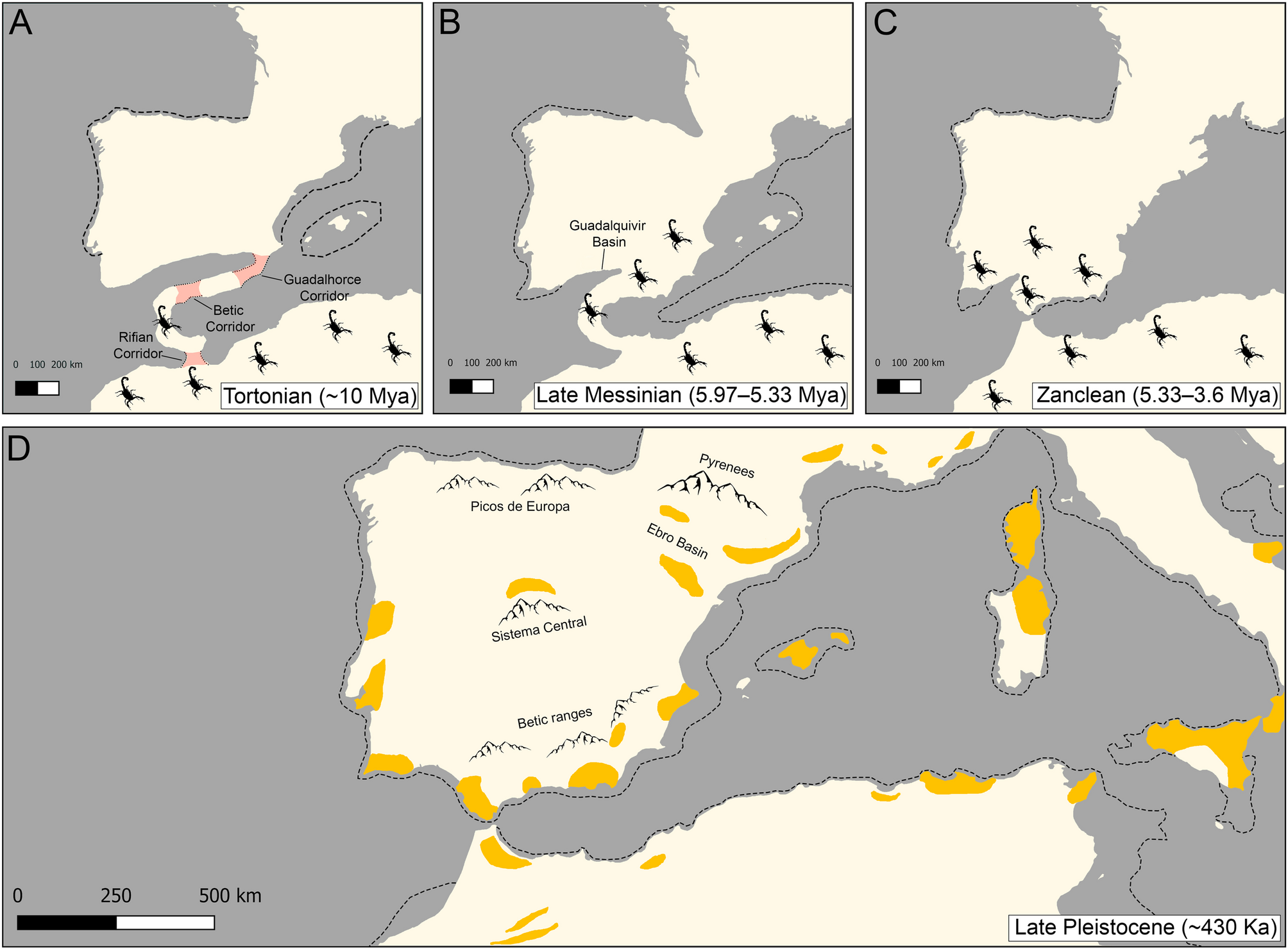
Toews DPL, Brelsford A. The biogeography of mitochondrial and nuclear discordance in animals. Mol Ecol. 2012;21:3907–30. https://doi.org/10.1111/j.1365-294X.2012.05664.x.
Google Scholar
Després L. One, two or more species? Mitonuclear discordance and species delimitation. Mol Ecol. 2019;28:3845–7. https://doi.org/10.1111/mec.15211.
Google Scholar
Wüster W, Kaiser H, Hoogmoed MS, Ceríaco LMP, Dirksen L, Dufresnes C, et al. How not to describe a species: lessons from a tangle of anacondas (Boidae: Eunectes Wagler, 1830). Zool J Linn Soc. 2024;201:zlae099. https://doi.org/10.1093/zoolinnean/zlae099.
Google Scholar
Funk DJ, Omland KE. Species-level paraphyly and polyphyly: frequency, causes, and consequences, with insights from animal mitochondrial DNA. Annu Rev Ecol Evol Syst. 2003;34:397–423. https://doi.org/10.1146/annurev.ecolsys.34.011802.132421.
Google Scholar
Ballard JWO, Whitlock MC. The incomplete natural history of mitochondria. Mol Ecol. 2004;13:729–44. https://doi.org/10.1046/j.1365-294X.2003.02063.x.
Google Scholar
Hurst GD, Jiggins FM. Problems with mitochondrial DNA as a marker in population, phylogeographic and phylogenetic studies: the effects of inherited symbionts. Proc R Soc Lond B Biol Sci. 2005;272:1525–34. https://doi.org/10.1098/rspb.2005.3056.
Google Scholar
Edwards SV, Potter S, Schmitt CJ, Bragg JG, Moritz C. Reticulation, divergence, and the phylogeography–phylogenetics continuum. Proc Natl Acad Sci U S A. 2016;113:8025–32. https://doi.org/10.1073/pnas.1601066113.
Google Scholar
Andújar C, Arribas P, Ruiz C, Serrano J, Gómez-Zurita J. Integration of conflict into integrative taxonomy: fitting hybridization in species delimitation of Mesocarabus (Coleoptera: Carabidae). Mol Ecol. 2014;23:4344–61. https://doi.org/10.1111/mec.12793.
Google Scholar
Firneno TJ, O’Neill JR, Itgen MW, Kihneman TA, Townsend JH, Fujita MK. Delimitation despite discordance: evaluating the species limits of a confounding species complex in the face of mitonuclear discordance. Ecol Evol. 2021;11:12739–53. https://doi.org/10.1002/ece3.8018.
Google Scholar
Abalde S, Crocetta F, Tenorio MJ, D’Aniello S, Fassio G, Rodríguez-Flores PC, Uribe JE, Afonso CML, Oliverio M, Zardoya R. Hidden species diversity and mito-nuclear discordance within the mediterranean cone snail, Lautoconus ventricosus. Mol Phylogenet Evol. 2023;186:107838. https://doi.org/10.1016/j.ympev.2023.107838.
Google Scholar
Duran DP, Laroche RA, Roman SJ, Godwin W, Herrmann DP, Bull E, et al. Species delimitation, discovery and conservation in a tiger beetle species complex despite discordant genetic data. Sci Rep. 2024;14:6617. https://doi.org/10.1038/s41598-024-56875-9.
Google Scholar
Carstens BC, Pelletier TA, Reid NM, Satler JD. How to fail at species delimitation. Mol Ecol. 2013;22:4369–83. https://doi.org/10.1111/mec.12413.
Google Scholar
Quattrini AM, Wu T, Soong K, Jeng M-S, Benayahu Y, McFadden CS. A next generation approach to species delimitation reveals the role of hybridization in a cryptic species complex of corals. BMC Evol Biol. 2019;19:116. https://doi.org/10.1186/s12862-019-1427-y.
Google Scholar
Dowling TE, Secor CL. The role of hybridization and introgression in the diversification of animals. Annu Rev Ecol Syst. 1997;28:593–619. https://doi.org/10.1146/annurev.ecolsys.28.1.593.
Google Scholar
Tovar-Sánchez E, Oyama K. Natural hybridization and hybrid zones between Quercus crassifolia and Quercus crassipes (Fagaceae) in Mexico: morphological and molecular evidence. Am J Bot. 2004;91:1352–63. https://doi.org/10.3732/ajb.91.9.1352.
Google Scholar
Renaud S, Alibert P, Auffray J-C. Modularity as a source of new morphological variation in the mandible of hybrid mice. BMC Evol Biol. 2012;12:141. https://doi.org/10.1186/1471-2148-12-141.
Google Scholar
Harvati K, Ackermann RR. Merging morphological and genetic evidence to assess hybridization in western Eurasian Late Pleistocene hominins. Nat Ecol Evol. 2022;6:1573–85. https://doi.org/10.1038/s41559-022-01875-z.
Google Scholar
Freitas I, Velo-Antón G, Kaliontzopoulou A, Zuazo Ó, Martínez-Freiría F. Association between genetic admixture and morphological patterns in a hybrid zone between the two Iberian vipers, Vipera aspis and V. latastei. J Zool Syst Evol Res. 2024;2024(3800363). https://doi.org/10.1155/2024/3800363.
Hewitt GM. Some genetic consequences of ice ages, and their role in divergence and speciation. Biol J Linn Soc. 1996;58:247–76. https://doi.org/10.1006/bijl.1996.0035.
Google Scholar
Hewitt G. The genetic legacy of the Quaternary ice ages. Nature. 2000;405:907–13. https://doi.org/10.1038/35016000.
Google Scholar
García-París M, Alcobendas M, Buckley D, Wake DB. Dispersal of viviparity across contact zones in Iberian populations of fire salamanders (Salamandra) inferred from discordance of genetic and morphological traits. Evolution. 2003;57:129–43. https://doi.org/10.1111/j.0014-3820.2003.tb00221.x.
Google Scholar
Schmitt T. Molecular biogeography of Europe: Pleistocene cycles and postglacial trends. Front Zool. 2007;4. https://doi.org/10.1186/1742-9994-4-11.
Bisconti R, Porretta D, Arduino P, Nascetti G, Canestrelli D. Hybridization and extensive mitochondrial introgression among fire salamanders in peninsular Italy. Sci Rep. 2018;8:13187. https://doi.org/10.1038/s41598-018-31535-x.
Dufresnes C, Pribille M, Alard B, Gonçalves H, Amat F, Crochet P-A, Dubey S, Perrin N, Fumagalli L, Vences M, Martínez-Solano I. Integrating hybrid zone analyses in species delimitation: lessons from two anuran radiations of the western Mediterranean. Heredity. 2020;124:423–38. https://doi.org/10.1038/s41437-020-0294-z.
Gómez, A, Lunt, DH. Refugia within refugia: patterns of phylogeographic concordance in the Iberian Peninsula. In: Weiss S, Ferrand N (eds), Phylogeography of southern European refugia. Springer, Dordrecht. https://doi.org/10.1007/1-4020-4904-8_5.
Paulo OS, Pinheiro J, Miraldo A, Bruford MW, Jordan WC, Nichols RA. The role of vicariance vs. dispersal in shaping genetic patterns in ocellated lizard species in the western Mediterranean. Mol Ecol. 2008;17:1535–51. https://doi.org/10.1111/j.1365-294X.2008.03706.x.
Médail F, Diadema K. Glacial refugia influence plant diversity patterns in the Mediterranean basin. J Biogeogr. 2009;36:1333–45. https://doi.org/10.1111/j.1365-2699.2008.02051.x.
Google Scholar
Costa GJ, Nunes VL, Marabuto E, Mendes R, Silva DN, Pons P, Bas JM, Hertach T, Paulo OS, Simões PC. The effect of the Messinian salinity crisis on the early diversification of the Tettigettalna cicadas. Zool Scr. 2023;52:100–16. https://doi.org/10.1111/zsc.12571.
Google Scholar
Gvoždík V, Canestrelli D, García-París M, Moravec J, Nascetti G, Recuero E, Teixeira J, Kotlík P. Speciation history and widespread introgression in the European short-call tree frogs (Hyla arborea sensu lato, H. intermedia and H. sarda). Mol Phylogenet Evol. 2015;83:143–55. https://doi.org/10.1016/j.ympev.2014.11.012.
Bassitta M, Buades JM, Pérez-Cembranos A, Pérez‐Mellado V, Terrasa B, Brown RP, Navarro P, Lluch J, Ortega J, Castro JA, Picornell A, Ramon C. Multilocus and morphological analysis of south‐eastern Iberian wall lizards (Squamata, Podarcis). Zool Scr. 2020;49:668–83. https://doi.org/10.1111/zsc.12450.
Google Scholar
Ambu J, Martínez-Solano Í, Suchan T, Hernandez A, Wielstra B, Crochet P-A, et al. Genomic phylogeography illuminates deep cyto-nuclear discordances in midwife toads (Alytes). Mol Phylogenet Evol. 2023;183:107783. https://doi.org/10.1016/j.ympev.2023.107783.
Google Scholar
Benson RH, Rakic-El Bied K, Bonaduce G. An important current reversal (influx) in the Rifian corridor (Morocco) at the Tortonian‐Messinian boundary: the end of Tethys Ocean. Paleoceanography. 1991;6:165–92. https://doi.org/10.1029/90PA00756.
Google Scholar
Sousa P, Harris DJ, Froufe E, van der Meijden A. Phylogeographic patterns of Buthus scorpions (Scorpiones: Buthidae) in the Maghreb and south-western Europe based on CO1 mtDNA sequences. J Zool. 2012;288:66–75. https://doi.org/10.1111/j.1469-7998.2012.00925.x.
Google Scholar
Sousa P. Resilient arthropods: Buthus scorpions as a model to understand the role of past and future climatic changes on Iberian biodiversity. Ph.D. Dissertation, Universidade do Porto (Portugal). 2017. https://repositorio-aberto.up.pt/handle/10216/106183.
Klesser R, Husemann M, Schmitt T, Sousa P, Moussi A, Habel JC. Molecular biogeography of the Mediterranean Buthus species complex (Scorpiones: Buthidae) at its southern Palaearctic margin. Biol J Linn Soc. 2021;133:166–78. https://doi.org/10.1093/biolinnean/blab014.
Google Scholar
Arntzen JW, García-París M. Morphological and allozyme studies of midwife toads (genus Alytes), including the description of two new taxa from Spain. Bijdr Dierkd. 1995;65:5–34. https://doi.org/10.1163/26660644-06501002.
Google Scholar
Kindler C, de Pous P, Carranza S, Beddek M, Geniez P, Fritz U. Phylogeography of the Ibero-Maghrebian red-eyed grass snake (Natrix astreptophora). Org Divers Evol. 2018;18:143–50. https://doi.org/10.1007/s13127-017-0354-2.
Google Scholar
Gantenbein B, Largiadèr CR. The phylogeographic importance of the Strait of Gibraltar as a gene flow barrier in terrestrial arthropods: a case study with the scorpion Buthus occitanus as model organism. Mol Phylogenet Evol. 2003;28:119–30. https://doi.org/10.1016/S1055-7903(03)00031-9.
Google Scholar
Gantenbein B. The genetic population structure of Buthus occitanus (Scorpiones: Buthidae) across the Strait of Gibraltar: calibrating a molecular clock using nuclear allozyme variation. Biol J Linn Soc. 2004;81:519–34. https://doi.org/10.1111/j.1095-8312.2003.00295.x.
Google Scholar
Gantenbein B, Fet V, Gantenbein-Ritter IA, Balloux F. Evidence for recombination in scorpion mitochondrial DNA (Scorpiones: Buthidae). Proc R Soc B Biol Sci. 2005;272:697–704. https://doi.org/10.1098/rspb.2004.3017.
Google Scholar
Habel JC, Husemann M, Schmitt T, Zachos FE, Honnen A-C, Petersen B, et al. Microallopatry caused strong diversification in Buthus scorpions (Scorpiones: Buthidae) in the Atlas Mountains (NW Africa). PLoS One. 2012;7:e29403. https://doi.org/10.1371/journal.pone.0029403.
Google Scholar
Pedroso D, Sousa P, Harris DJ, Van der Meijden A. Phylogeography of Buthus Leach, 1815 (Scorpiones: Buthidae): a multigene molecular approach reveals a further complex evolutionary history in the Maghreb. Afr Zool. 2013;48:298–308. https://doi.org/10.3377/004.048.0216.
Google Scholar
Blasco-Aróstegui J, Simone Y, Prendini L. Systematic revision of the European species of Buthus Leach, 1815 (Scorpiones: Buthidae). Bull Am Mus Nat Hist. 2025;476:1–131. https://doi.org/10.1206/0003-0090.476.1.1.
Sánchez-Piñero F, Urbano-Tenorio F. Watch out for your neighbor: climbing onto shrubs is related to risk of cannibalism in the scorpion Buthus cf. occitanus. PLoS One. 2016;11:e0161747. https://doi.org/10.1371/journal.pone.0161747.
Google Scholar
Sánchez-Piñero F, Urbano-Tenorio F, Puerta-Rodríguez L. Foraging strategies, prey selection and size- and microhabitat-related diet variation in Buthus montanus (Scorpiones: Buthidae) in an arid area of SE Spain. J Arachnol. 2025;52:189–98. https://doi.org/10.1636/JoA-S-23-008.
Google Scholar
Tuzet O. Sur La spermatogénèse de Buthus occitanus. Arch Zool Expérimentale Générale. 1938;80:335–51.
Sloan DB, Havird JC, Sharbrough J. The on-again, off‐again relationship between mitochondrial genomes and species boundaries. Mol Ecol. 2017;26:2212–36. https://doi.org/10.1111/mec.13959.
Google Scholar
Teruel R, Turiel C. The genus Buthus Leach, 1815 (Scorpiones: Buthidae) in the Iberian Peninsula. Part 5: A new psammophile species from northern Spain, a synonymy and first albinism record in the genus. Rev Ibérica Aracnol. 2022;41:15–36.
Ythier E. The southernmost scorpion species in Europe: Buthus gabani sp. n. from Cape St. Vincent, Algarve, Portugal (Scorpiones: Buthidae). Faunitaxys. 2021;9:1–6.
Ythier E. The genus Buthus Leach, 1815 (Scorpiones: Buthidae) in France with description of a new species from the eastern Pyrenees. Faunitaxys. 2021;9:1–10.
Ythier E, Laborieux L. The genus Buthus Leach, 1815 (Scorpiones: Buthidae) in France with description of a new species from the Sainte-Baume Massif. Faunitaxys. 2022;10:1–13.
González-Moliné A, Armas, de LF. Una especie nueva del género Buthus (Scorpiones: Buthidae) de la Provincia de Huelva, España. Rev Ibérica Arachnol. 2024;44:75–84.
Sousa P, Froufe E, Alves PC, Harris DJ. Genetic diversity within scorpions of the genus Buthus from the Iberian Peninsula: mitochondrial DNA sequence data indicate additional distinct cryptic lineages. J Arachnol. 2010;38:206–11.
Google Scholar
Vachon M. Études sur le scorpions. Arch l’Institut Pasteur d’Algérie. 1952;1–482.
Lourenço WR, Vachon M. Considérations sur le genre Buthus Leach, 1815 en Espagne, et description de deux nouvelles espèces (Scorpiones, Buthidae), Rev. Ibérica Aracnol. 2004;9:81–4.
Lourenço WR. Une nouvelle espèce appartenant au genre Buthus Leach, 1815 (Scorpiones: Buthidae) collectée dans le Parc Naturel de la ‘Serra da Estrela’ au centre du Portugal. Faunitaxys. 2021;9:1–7.
Rossi A. Notes on the distribution of the species of the genus Buthus (Leach, 1815) (Scorpiones, Buthidae) in Europe, with a description of a new species from Spain. Bull Br Arachnol Soc. 2012;15:273–9.
Teruel R, Turiel C. The genus Buthus Leach, 1815 (Scorpiones: Buthidae) in the Iberian Peninsula. Part 1: four redescriptions and six new species. Rev Ibérica Arachnol. 2020;37:3–60.
Teruel R, Turiel C. The genus Buthus Leach, 1815 (Scorpiones: Buthidae) in the Iberian Peninsula. Part 2: two more redescriptions. Rev Ibérica Aracnol. 2021;38:3–20.
Teruel R, Turiel C. The genus Buthus Leach, 1815 (Scorpiones: Buthidae) in the Iberian Peninsula. Part 3: a new species from central Spain and new records. Rev Ibérica Aracnol. 2021;39:3–11.
Teruel R, Turiel C. The genus Buthus Leach, 1815 (Scorpiones: Buthidae) in the Iberian Peninsula. Part 4: a new species from southern Spain. Rev Ibérica Aracnol. 2022;40:19–29.
Sousa P, Arnedo M, Harris DJ. Updated catalogue and taxonomic notes on the Old-World scorpion genus Buthus Leach, 1815 (Scorpiones, Buthidae). ZooKeys. 2017;686:15–84. https://doi.org/10.3897/zookeys.686.12206.
Google Scholar
Maddison WP, Maddison DR. Mesquite: a modular system for evolutionary analysis. Version 3.81, (2023) http://www.mesquiteproject.org
Goloboff PA, Mattoni CI, Quinteros AS. Continuous characters analyzed as such. Cladistics. 2006;22:589–601. https://doi.org/10.1111/j.1096-0031.2006.00122.x.
Google Scholar
Hwang U-W, Kim W. General properties and phylogenetic utilities of nuclear ribosomal DNA and mitochondrial DNA commonly used in molecular systematics. Korean J Parasitol. 1999;37:215. https://doi.org/10.3347/kjp.1999.37.4.215.
Google Scholar
Bryson RW, Riddle BR, Graham MR, Smith BT, Prendini L. As old as the hills: montane scorpions in southwestern North America reveal ancient associations between biotic diversification and landscape history. PLoS ONE. 2013;8:e52822. https://doi.org/10.1371/journal.pone.0052822.
Google Scholar
Cain S, Loria SF, Ben-Shlomo R, Prendini L, Gefen E. Dated phylogeny and ancestral range estimation of sand scorpions (Buthidae: Buthacus) reveal Early Miocene divergence across land bridges connecting Africa and Asia. Mol Phylogenet Evol. 2021;164. https://doi.org/10.1016/j.ympev.2021.107212.
Loria SF, Prendini L. Burrowing into the forest: phylogeny of the Asian forest scorpions (Scorpionidae: Heterometrinae) and the evolution of ecomorphotypes. Cladistics. 2021;37:109–61. https://doi.org/10.1111/cla.12434.
Google Scholar
Loria SF, Ehrenthal VL, Nguyen AD, Prendini L. Climate relicts: Asian scorpion family Pseudochactidae survived Miocene aridification in caves of the Annamite Mountains. Insect Syst Divers. 2022;3:1–21. https://doi.org/10.1093/isd/ixac028.
Katoh K. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 2002;30:3059–66. https://doi.org/10.1093/nar/gkf436.
Google Scholar
Katoh K, Standley DM. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 2013;30:772–80. https://doi.org/10.1093/molbev/mst010.
Google Scholar
Katoh K, Toh H. Recent developments in the MAFFT multiple sequence alignment program. Brief Bioinform. 2008;9:286–98. https://doi.org/10.1093/bib/bbn013.
Google Scholar
Vaidya G, Lohman DJ, Meier R. SequenceMatrix: concatenation software for the fast assembly of multi-gene datasets with character set and codon information. Cladistics. 2011;27:171–80. https://doi.org/10.1111/j.1096-0031.2010.00329.x.
Google Scholar
Tamura K, Stecher G, Kumar S. MEGA11. Molecular evolutionary genetics analysis version 11. Mol. Biol Evol. 2021;38:3022–7. https://doi.org/10.1093/molbev/msab120.
Google Scholar
Minh BQ, Schmidt HA, Chernomor O, Schrempf D, Woodhams MD, Von Haeseler A, et al. IQ-TREE 2: new models and efficient methods for phylogenetic inference in the genomic era. Mol Biol Evol. 2020;37:1530–4. https://doi.org/10.1093/molbev/msaa015.
Ronquist F, Huelsenbeck JP. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:1572–4. https://doi.org/10.1093/bioinformatics/btg180.
Ronquist F, Teslenko M, Van der Mark P, Ayres DL, Darling A, Höhna S, et al. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 2012;61:539–42. https://doi.org/10.1093/sysbio/sys029.
Google Scholar
Bianchini G, Sánchez-Baracaldo P. TreeViewer: flexible, modular software to visualise and manipulate phylogenetic trees. Ecol Evol. 2024;14:e10873. https://doi.org/10.1002/ece3.10873.
Google Scholar
Kalyaanamoorthy S, Minh BQ, Wong TKF, Von Haeseler A, Jermiin LS. ModelFinder: fast model selection for accurate phylogenetic estimates. Nat Methods. 2017;14:587–9. https://doi.org/10.1038/nmeth.4285.
Google Scholar
Huelsenbeck JP. Bayesian phylogenetic model selection using reversible jump Markov chain Monte Carlo. Mol Biol Evol. 2004;21:1123–33. https://doi.org/10.1093/molbev/msh123.
Google Scholar
Lewis PO. A likelihood approach to estimating phylogeny from discrete morphological character data. Syst Biol. 2001;50:913–25. https://doi.org/10.1080/106351501753462876.
Google Scholar
Nixon KC, Carpenter JM. On simultaneous analysis. Cladistics. 1996;12:221–41. https://doi.org/10.1111/j.1096-0031.1996.tb00010.x.
Google Scholar
Prendini L, Crowe TM, Wheeler WC. Systematics and biogeography of the family Scorpionidae (Chelicerata: Scorpiones), with a discussion on phylogenetic methods. Invertebr Syst. 2003;17:185. https://doi.org/10.1071/IS02016.
Google Scholar
De Sá RO, Grant T, Camargo A, Heyer WR, Ponssa ML, Stanley E. Systematics of the Neotropical genus Leptodactylus fitzinger, 1826 (Anura: Leptodactylidae): phylogeny, the relevance of non-molecular evidence, and species accounts. South Am J Herpetol. 2014;S100. https://doi.org/10.2994/SAJH-D-13-00022.1.
Wilson JD, Raven RJ, Schmidt DJ, Hughes JM, Rix MG. Total-evidence analysis of an undescribed fauna: resolving the evolution and classification of Australia’s golden trapdoor spiders (Idiopidae: Arbanitinae: Euoplini). Cladistics. 2020;36:543–68. https://doi.org/10.1111/cla.12415.
Google Scholar
Cain S, Gefen E, Prendini L. Systematic revision of the sand scorpions, genus Buthacus Birula, 1908 (Buthidae C.L. Koch, 1837) of the Levant, with redescription of Buthacus arenicola (Simon, 1885) from Algeria and Tunisia. Bull Am Mus Nat Hist. 2021;450(1):1–136. https://doi.org/10.1206/0003-0090.450.1.1.
Google Scholar
Minh BQ, Nguyen MAT, Von Haeseler A. Ultrafast approximation for phylogenetic bootstrap. Mol Biol Evol. 2013;30:1188–95. https://doi.org/10.1093/molbev/mst024.
Google Scholar
Hoang DT, Chernomor O, Von Haeseler A, Minh BQ, Vinh LS. UFBoot2: improving the ultrafast bootstrap approximation. Mol Biol Evol. 2018;35:518–22. https://doi.org/10.1093/molbev/msx281.
Google Scholar
Sankoff D, Abel Y, Hein J. A tree, a window, a hill; generalization of nearest-neighbor interchange in phylogenetic optimization. J Classif. 1994;11:209–32. https://doi.org/10.1007/BF01195680.
Google Scholar
Guindon S, Dufayard J-F, Lefort V, Anisimova M, Hordijk W, Gascuel O. New algorithms and methods to estimate Maximum-Likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol. 2010;59:307–21. https://doi.org/10.1093/sysbio/syq010.
Google Scholar
Huson DH, Bryant D. Application of phylogenetic networks in evolutionary studies. Mol Biol Evol. 2006;23:254–67. https://doi.org/10.1093/molbev/msj030.
Google Scholar
Huson DH, Bryant D. The SplitsTree app: interactive analysis and visualization using phylogenetic trees and networks. Nat Methods. 2024;21:1773–4. https://doi.org/10.1038/s41592-024-02406-3.
Google Scholar
Zhang L, Abhari N, Colijn C, Wu Y. A fast and scalable method for inferring phylogenetic networks from trees by aligning lineage taxon strings. Genome Res. 2023;33:1053–60. https://doi.org/10.1101/gr.277669.123.
Google Scholar
Zhang L, Cetinkaya B, Huson DH. PhyloFusion—fast and easy fusion of rooted phylogenetic trees into rooted phylogenetic networks. Syst Biol. 2025. https://doi.org/10.1093/sysbio/syaf049.
Google Scholar
Venables WN, Ripley BD. Modern applied statistics with S. 4th ed. Springer, New York; 2002.
Google Scholar
Kuhn M. Building predictive models in R using the caret package. J Stat Softw. 2008;28:1–26. https://doi.org/10.18637/jss.v028.i05.
Google Scholar
Raxworthy CJ, Ingram CM, Rabibisoa N, Pearson RG. Applications of ecological niche modeling for species delimitation: a review and empirical evaluation using day geckos (Phelsuma) from Madagascar. Syst Biol. 2007;56:907–23. https://doi.org/10.1080/10635150701775111.
Google Scholar
Goodman A, Allen J, Brim J, Codella A, Hahn B, Jojo H, BondocGawa Mafla-Mills S, Bondoc Mafla ST, Oduro A, Wilson M, Ware, J. Utilization of community science data to explore habitat suitability of basal termite genera. Insect Syst Divers. 2022;6:1–15. https://doi.org/10.1093/isd/ixac019.
Google Scholar
Fick SE, Hijmans RJ. Worldclim 2: new 1-km spatial resolution climate surfaces for global land areas. Int J Climatol. 2017;37:4302–15. https://doi.org/10.1002/joc.5086.
Google Scholar
Moo-Llanes DA, López-Ordóñez T, Torres-Monzón JA, Mosso-González C, Casas-Martínez M, Samy AM. Assessing the potential distributions of the invasive mosquito vector Aedes albopictus and its natural Wolbachia infections in México. Insects. 2021;12:143. https://doi.org/10.3390/insects12020143.
Google Scholar
Prendini L. Substratum specialization and speciation in southern African scorpions: the Effect Hypothesis revisited. In: Fet V, Seiden P, editors. Scorpions 2001. In Memoriam Gary A. Polis.: British Arachnological Society, Burnham Beeches, UK; 2001. p. 113–38.
Hengl T, Mendes de Jesus J, Heuvelink GBM, Ruiperez Gonzalez M, Kilibarda M, Blagotićć A, et al. SoilGrids250m: global gridded soil information based on machine learning. PLoS One. 2017;12:e0169748. https://doi.org/10.1371/journal.pone.0169748.
Brun P, Zimmermann NE, Hari C, Pellissier L, Karger DN. Global climate-related predictors at kilometer resolution for the past and future. Earth Syst Sci Data. 2022;14:5573–603. https://doi.org/10.5194/essd-14-5573-2022.
Google Scholar
Phillips SJ, Anderson RP, Schapire RE. Maximum entropy modeling of species geographic distributions. Ecol Modell. 2006;190:231–59. https://doi.org/10.1016/j.ecolmodel.2005.03.026.
Google Scholar
Calatayud-Mascarell A, Alonso-Alonso P, Boratyński Z, Dippenaar-Schoeman A, Pabijan M, Salgado-Irazabal X. Hidden among the prickles: new records and updated distribution of Tmarus longicaudatus Millot, 1942 (Araneae: Thomisidae). Arachnology. 2022;19:1–6. https://doi.org/10.13156/arac.2022.19.1.1.
Sánchez‐Vialas A, Calatayud‐Mascarell A, Recuero E, Ruiz JL, García‐París M. Predictions based on phylogeography and climatic niche modelling depict an uncertain future scenario for giant blister beetles (Berberomeloe, Meloidae) facing intensive greenhouse expansion and global warming. Insect Conserv Divers. 2023;16:801–16. https://doi.org/10.1111/icad.12671.
Google Scholar
Peterson AT, Soberón J, Pearson RG, Anderson RP, Martínez-Meyer E, Nakamura M, Araújo MB. Ecological niches and geographic distributions. Princeton University Press, Princeton; 2011.
Phillips S. A brief tutorial on Maxent. Lessons Conserv. 2010;3:108–35. https://doi.org/10.5531/cbc.linc.3.1.6.
Google Scholar
Searcy CA, Shaffer HB. Do ecological niche models accurately identify climatic determinants of species ranges? Am Nat. 2016;187:423–35. https://doi.org/10.1086/685387.
Google Scholar
Wright K, Wright A. corrgram: plot a correlogram. Version 1.8 [R package], (2018) https://cran.r-project.org/web/packages/corrgram/i.
Dormann CF, Elith J, Bacher S, Buchmann C, Carl G, Carré G, et al. Collinearity: a review of methods to deal with it and a simulation study evaluating their performance. Ecography. 2013;36:27–46. https://doi.org/10.1111/j.1600-0587.2012.07348.x.
Google Scholar
Harrell FE. Regression modeling strategies: With applications to linear models, logistic and ordinal regression, and survival analysis, 2nd ed., Springer International Publishing, Cham. 2015. https://doi.org/10.1007/978-3-319-19425-7
Polidori C, García-Gila J, Blasco-Aróstegui J, Gil-Tapetado D. Urban areas are favouring the spread of an alien mud-dauber wasp into climatically non-optimal latitudes. Acta Oecol. 2021;110:103678. https://doi.org/10.1016/j.actao.2020.103678.
Google Scholar
Gómez SR, Gil-Tapetado D, García‐Gila J, Blasco‐Aróstegui J, Polidori C. The leaf beetle Labidostomis lusitanica (Coleoptera: Chrysomelidae) as an Iberian pistachio pest: projecting risky areas. Pest Manag Sci. 2022;78:217–29. https://doi.org/10.1002/ps.6624.
Google Scholar
Oksanen J, Simpson GL, Blanchet FG, Kindt R, Legendre P, Minchin PR, et al. vegan: community ecology package. R package version 2.6–10; 2022. https://CRAN.R-project.org/package=vegan.
Goodman A, Esposito L. Niche partitioning in congeneric scorpions. Invertebr Biol. 2020;139:e12280. https://doi.org/10.1111/ivb.12280.
Google Scholar
Warren DL, Matzke NJ, Cardillo M, Baumgartner JB, Beaumont LJ, Turelli M, et al. ENMtools 1.0: an R package for comparative ecological biogeography. Ecography. 2021;44:504–11. https://doi.org/10.1111/ecog.05485.
Google Scholar
Wooten JA, Gibbs HL. Niche divergence and lineage diversification among closely related Sistrurus rattlesnakes. J Evol Biol. 2012;25:317–28. https://doi.org/10.1111/j.1420-9101.2011.02426.x.
Google Scholar
Van der Vaart AW. Asymptotic statistics. Cambridge University Press, Cambridge, U.K.; 1998. https://doi.org/10.1017/CBO9780511802256
Warren DL, Glor RE, Turelli M. Environmental niche equivalency versus conservatism: quantitative approaches to niche evolution. Evolution. 2008;62:2868–83. https://doi.org/10.1111/j.1558-5646.2008.00482.x.
Google Scholar
Barahoei H, Prendini L, Navidpour S, Tahir HM, Aliabadian M, Siahsarvie R, et al. Integrative systematics of the tooth-tailed scorpions, Odontobuthus (Buthidae), with descriptions of three new species from the Iranian Plateau. Zool J Linn Soc. 2022;195:355–98. https://doi.org/10.1093/zoolinnean/zlab030.
Google Scholar
Amiri M, Prendini L, Hussen FS, Aliabadian M, Siahsarvie R, Mirshamsi O. Integrative systematics of the widespread Middle Eastern buthid scorpion, Hottentotta saulcyi (Simon, 1880), reveals a new species in Iran. Arthropod Syst Phylogeny. 2024;82:323–41. https://doi.org/10.3897/asp.82.e98662.
Google Scholar
Bergsten J. A review of long-branch attraction. Cladistics. 2005;21:163–93. https://doi.org/10.1111/j.1096-0031.2005.00059.x.
Google Scholar
Yang Z, Zhu T. Bayesian selection of misspecified models is overconfident and may cause spurious posterior probabilities for phylogenetic trees. Proc Natl Acad Sci U S A. 2018;115:1854–9. https://doi.org/10.1073/pnas.1712673115.
Google Scholar
Douady CJ, Delsuc F, Boucher Y, Doolittle WF, Douzery EJP. Comparison of Bayesian and Maximum Likelihood bootstrap measures of phylogenetic reliability. Mol Biol Evol. 2003;20:248–54. https://doi.org/10.1093/molbev/msg042.
Google Scholar
Erixon P, Svennblad B, Britton T, Oxelman B. Reliability of Bayesian posterior probabilities and bootstrap frequencies in phylogenetics. Syst Biol. 2003;52:665–73. https://doi.org/10.1080/10635150390235485.
Google Scholar
Irwin DE. Phylogeographic breaks without geographic barriers to gene flow. Evolution. 2002;56:2383–94. https://doi.org/10.1111/j.0014-3820.2002.tb00164.x.
Google Scholar
Avise JC. Molecular markers, natural history and evolution. Springer, New York; 1994.
Gantenbein B, Largiadèr CR. Mesobuthus gibbosus (Scorpiones: Buthidae) on the island of Rhodes — hybridization between Ulysses’ stowaways and native scorpions? Mol Ecol. 2002;11:925–38. https://doi.org/10.1046/j.1365-294X.2002.01494.x.
Google Scholar
Kunerth HD, Tapisso JT, Valente R, Mathias MdaL, Alves PC, Searle JB, et al. Characterising mitochondrial capture in an Iberian shrew. Genes. 2022;13:2228. https://doi.org/10.3390/genes13122228.
Google Scholar
Due D, Polis GA. The biology of Vaejovis littoralis Williams, an intertidal scorpion from Baja California, Mexico. J Zool. 1985;207:563–80. https://doi.org/10.1111/j.1469-7998.1985.tb04952.x.
Starrett J, Hedin M. Multilocus genealogies reveal multiple cryptic species and biogeographical complexity in the California turret spider Antrodiaetus riversi (Mygalomorphae, Antrodiaetidae). Mol Ecol. 2007;16:583–604. https://doi.org/10.1111/j.1365-294X.2006.03164.x.
Google Scholar
Kaltsas D, Mylonas M. Locomotory activity and orientation of Mesobuthus gibbosus (Scorpiones: Buthidae) in central Aegean Archipelago. J Nat Hist. 2010;44:1445–59. https://doi.org/10.1080/00222931003632732.
Google Scholar
Hedin M, Starrett J, Hayashi C. Crossing the uncrossable: novel trans-valley biogeographic patterns revealed in the genetic history of low-dispersal mygalomorph spiders (Antrodiaetidae, Antrodiaetus) from California. Mol Ecol. 2013;22:508–26. https://doi.org/10.1111/mec.12130.
Google Scholar
Hill GE, Johnson JD. The mitonuclear compatibility hypothesis of sexual selection. Proc Biol Sci. 2013;280:20131314. https://doi.org/10.1098/rspb.2013.1314.
Google Scholar
Avise J, Shapira J, Daniel S, Aquadro C, Lansman R. Mitochondrial DNA differentiation during the speciation process in Peromyscus. Mol Biol Evol. 1984. https://doi.org/10.1093/oxfordjournals.molbev.a040301.
Google Scholar
Pamilo P, Nei M. Relationships between gene trees and species trees. Mol Biol Evol. 1988;5:568–83. https://doi.org/10.1093/oxfordjournals.molbev.a040517.
Google Scholar
Maddison WP, Knowles LL. Inferring phylogeny despite incomplete lineage sorting. Syst Biol. 2006;55:21–30. https://doi.org/10.1080/10635150500354928.
Google Scholar
Mas-Peinado P, García-París M, Ruiz JL, Buckley D. The Strait of Gibraltar is an ineffective palaeogeographic barrier for some flightless darkling beetles (Coleoptera: Tenebrionidae: Pimelia). Zool J Linn Soc. 2022;195:1147–80. https://doi.org/10.1093/zoolinnean/zlab088.
Google Scholar
Wirtz P. Mother species–father species: unidirectional hybridization in animals with female choice. Anim Behav. 1999;58:1–12. https://doi.org/10.1006/anbe.1999.1144.
Google Scholar
Polis GA. The biology of scorpions. Stanford University, Stanford, CA; 1990.
McLean CJ, Garwood RJ, Brassey CA. Sexual dimorphism in the arachnid orders. PeerJ. 2018;6:e5751. https://doi.org/10.7717/peerj.5751.
Sullivan JP, Lavoué S, Arnegard ME, Hopkins CD. AFLPs resolve phylogeny and reveal mitochondrial introgression within a species flock of African electric fish (Mormyroidea: Teleostei). Evolution. 2004;58:825–41. https://doi.org/10.1111/j.0014-3820.2004.tb00415.x.
Google Scholar
Papakostas S, Michaloudi E, Proios K, Brehm M, Verhage L, Rota J, et al. Integrative taxonomy recognizes evolutionary units despite widespread mitonuclear discordance: evidence from a rotifer cryptic species complex. Syst Biol. 2016;65:508–24. https://doi.org/10.1093/sysbio/syw016.
Google Scholar
Nahon D, Trompette R. Origin of siltstones: glacial grinding versus weathering. Sedimentology. 1982;29:25–35. https://doi.org/10.1111/j.1365-3091.1982.tb01706.x.
Google Scholar
Vrba ES. Evolution, species and fossils: how does life evolve? S Afr J Sci. 1980;76:61–84.
Raszick TJ, Song H. The ecotype paradigm: testing the concept in an ecologically divergent grasshopper. Insect Syst Evol. 2016;47:363–87. https://doi.org/10.1163/1876312X-47032147.
Google Scholar
Jiménez-Hernández VS, Villegas-Guzmán GA, Casasola-González JA, Vargas-Mendoza CF. Altitudinal distribution of alpha, beta, and gamma diversity of pseudoscorpions (Arachnida) in Oaxaca, Mexico. Acta Oecol. 2020;103:103525. https://doi.org/10.1016/j.actao.2020.103525.
Google Scholar
Blasco-Aróstegui J, Prendini L. Glacial relicts? A new scorpion from Mount Olympus, Greece (Euscorpiidae: Euscorpius). Am Mus Novit. 2023;4003:1–36. https://doi.org/10.1206/4003.1
Google Scholar
Lobo JM, Martín-Piera F. Searching for a predictive model for species richness of Iberian dung beetle based on spatial and environmental variables. Conserv Biol. 2002;16:158–73. https://doi.org/10.1046/j.1523-1739.2002.00211.x.
Google Scholar
Soberón J, Arroyo-Peña B. Are fundamental niches larger than the realized? Testing a 50-year-old prediction by Hutchinson. PLoS ONE. 2017;12:e0175138. https://doi.org/10.1371/journal.pone.0175138.
Google Scholar
Multigner LF, Gil-Tapetado D, Nieves-Aldrey JL, Gómez JF. The paths of the galls: differences in the ecology and distribution of two European oak gall wasps Andricus dentimitratus and Andricus pictus. J Zool Syst Evol Res. 2022;2022:1–14. https://doi.org/10.1155/2022/8488412.
Google Scholar

Sydney, 23 October 2025: Partner appointment will further bolster the firm’s corporate insurance and regulatory offering in the Asia Pacific region.
Global law firm Clyde & Co today announces the appointment of Yvonne Lam as partner. She re-joins the firm from Gilchrist Connell, where she was a Principal and the Head of Corporate Insurance & Insurtech, after previously spending over a decade specialising in advising on corporate insurance and regulatory matters at Clyde & Co in Australia.
Yvonne will further grow the firm’s corporate insurance offering in the Asia Pacific region, with a focus on handling transactional, regulatory and commercial advisory matters for clients in the rapidly evolving insurance risk and regulatory environment across Australia and Singapore.
Yvonne brings extensive experience in corporate insurance and regulatory matters and has advised clients on matters in the Australian insurance industry, including the Financial Services Royal Commission hearings and significant transactions arising from global mergers and acquisitions, corporate restructures and expansions into the Australian market. She acts for insurers and reinsurers, underwriting agents, insurance brokers and next-generation insurtech clients and fintech ventures in the Asia Pacific region.
She has been a finalist for the Lawyers Weekly New Partner of the Year Awards (Big Law, 2025), Women in Law Awards for Partner of the Year (Big Law, 2024) and Special Counsel of the Year (2021) and the Lawyers Weekly 30 Under 30 Awards for Commercial Law (2018). She is the Asian Australian Lawyers Association NSW Branch President for 2025 and has been recognised as a finalist in the Legal & Professionals category of the Asian Australian Leadership Awards in 2024.
Rebecca Kelly, Australia Managing Partner, Brisbane, said: “I’m delighted to welcome Yvonne back to the firm. Her return marks an exciting step for the firm in the continued growth of our corporate insurance and regulatory practice across Australia and the broader Asia Pacific region. With our enhanced capabilities, we are strongly positioned to support our clients in managing risk, responding to regulatory change, and navigating the complexities of today’s corporate landscape. Our clients rely on us for clear, commercially focused advice, and this appointment ensures we continue to deliver with confidence and clarity.”
Gareth Horne, Head of Insurance, Australia, Sydney, said: “This growth will allow us to seize the opportunities arising from the rapidly changing regulatory environment, where we see strong client demand for help with navigating the evolving risk landscape.”
Yvonne Lam said: “I am pleased to return to Clyde & Co and draw on the firm’s global platform and international reach particularly for my clients in the Asia Pacific region, who operate across multiple jurisdictions and in complex regulatory environments.”
The appointment further strengthens Clyde & Co’s regional transactional, corporate and regulatory capabilities servicing insurance clients across Australia and the wider Asia Pacific region.
Clyde & Co’s global insurance practice of 2,400 lawyers in over 70 offices around the world handles matters across all lines of insurance business, helping the insurance market navigate risk.
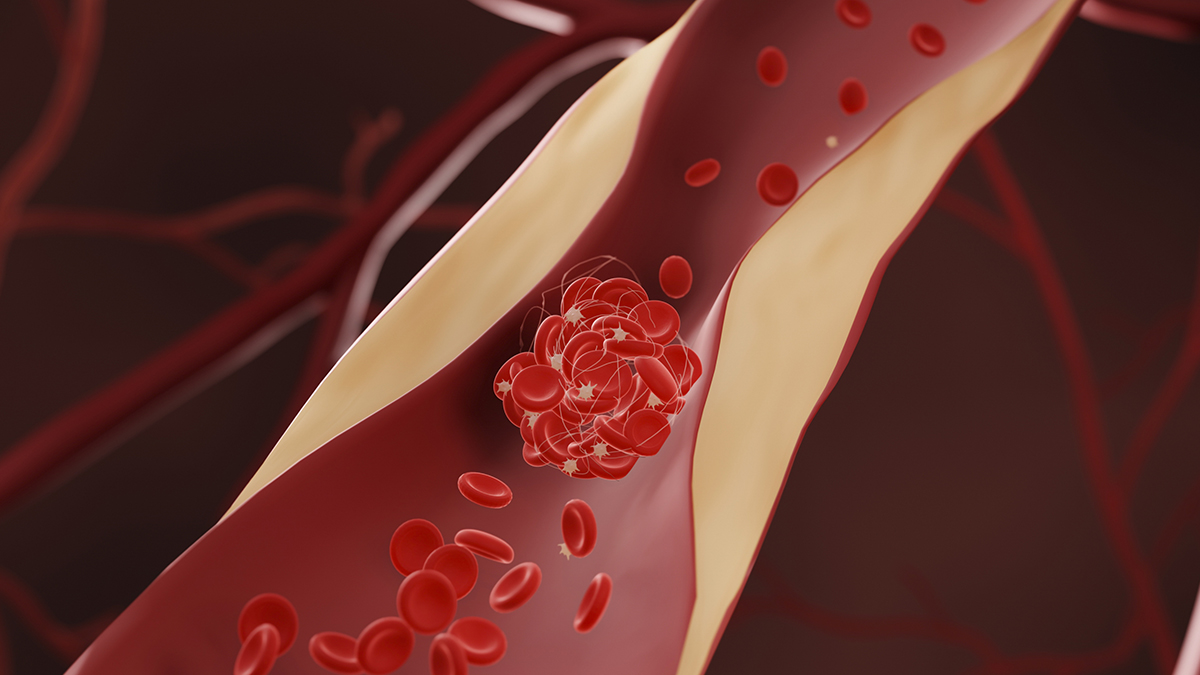
Reducing cholesterol levels might not just be good for your heart; they might also be effective at reducing your risk of dementia, according to a comprehensive meta-analysis involving close to a million participants.
The results imply that…

Newly appointed Japanese Prime Minister Sanae Takaichi started off her term with high approval ratings, in a show of optimism that she could deliver on her pledges that include countering inflation and strengthening the economy.
Takaichi has…

Cancer is one of the deadliest diseases worldwide and remains the second-leading cause of death. Despite major progress in medical treatments, early detection, and preventive care, it continues to be a serious global health concern. Experts…
BEIJING, Oct. 21 — In 1985, the eastern Chinese city of Xiamen was a study in contrasts.
A newly designated Special Economic Zone, it buzzed with the anxious energy of a nation just cracking…