By this point in October there is no escaping the shorter days, lower light and autumn drizzle, but this concert by London Symphony Orchestra – the first in a short series of LSO programmes this season and next curated by the composer and…
Blog
-
Just a moment…
Just a moment… This request seems a bit unusual, so we need to confirm that you’re human. Please press and hold the button until it turns completely green. Thank you for your cooperation!
Continue Reading
-

Single-Particle ICP-TOF-MS Enables High-Throughput Isotopic Analysis of Uranium Particles for Nuclear Safeguards
Since the 1970s, the International Atomic Energy Agency (IAEA) has relied on environmental sampling to verify nuclear materials, traditionally using bulk digestion and particle-level mass spectrometry methods such as thermal ionization mass…
Continue Reading
-

Assessing High-Dose Versus Standard-Dose Influenza Vaccine Protection Against Severe Disease Outcomes
This infographic summarizes findings from FLUNITY-HD, the largest ever individually randomized influenza vaccine effectiveness trial. Conducted across nearly half a million older adults, the study provides valuable real-world insights into the…
Continue Reading
-
SAP Named a Leader in the 2025 Gartner® Magic Quadrant™ for Cloud ERP for Product-Centric Enterprises – SAP News Center
- SAP Named a Leader in the 2025 Gartner® Magic Quadrant™ for Cloud ERP for Product-Centric Enterprises SAP News Center
- How Workday’s (WDAY) Expanded Partnership and Gartner Recognition Shape Its Higher Ed Investment Case simplywall.st
- Workday Recognized as a Leader in 2025 Gartner® Magic Quadrant™ for Cloud ERP for Service-Centric Enterprises for Fourth Consecutive Year Workday
- Workday’s Higher Education Momentum Accelerates as Over 650 Institutions Worldwide Turn to AI to Navigate Uncertainty and Transform the Student Experience Yahoo Finance
- 650+ Institutions Choose Workday for AI Campus Tools Worldwide; Workday Student Supports 3M Records Stock Titan
Continue Reading
-
Commitment to Privacy – Virginia Commonwealth University
We collect limited information about web visitors and use cookies on our website to provide you with the most optimal experience. These cookies help us provide you with personalized content and improve our website. To learn more about our web…
Continue Reading
-
Just a moment…
Just a moment… This request seems a bit unusual, so we need to confirm that you’re human. Please press and hold the button until it turns completely green. Thank you for your cooperation!
Continue Reading
-

Soybean farmers have another biological tool in the fight against pest
Ag input company Indigo Ag is helping soybean farmers address one of their largest pest problems ─ soybean cyst nematode (SCN) ─ with a biological that first launched in Brazil but is now making its way to the U.S. with Environmental…
Continue Reading
-
Champions League Matchday 3 possible line-ups and team news – UEFA.com
- Champions League Matchday 3 possible line-ups and team news UEFA.com
- UEFA Champions League Matchday 3 storylines, schedule, live stream: How to watch UCL on Paramount+ CBS Sports
- Predicted Line-Ups for Champions League Matchday 3 (2025/26) –…
Continue Reading
-
Development of a radiomics-3D deep learning fusion model for prognostic prediction in pancreatic cancer | BMC Cancer
Patients
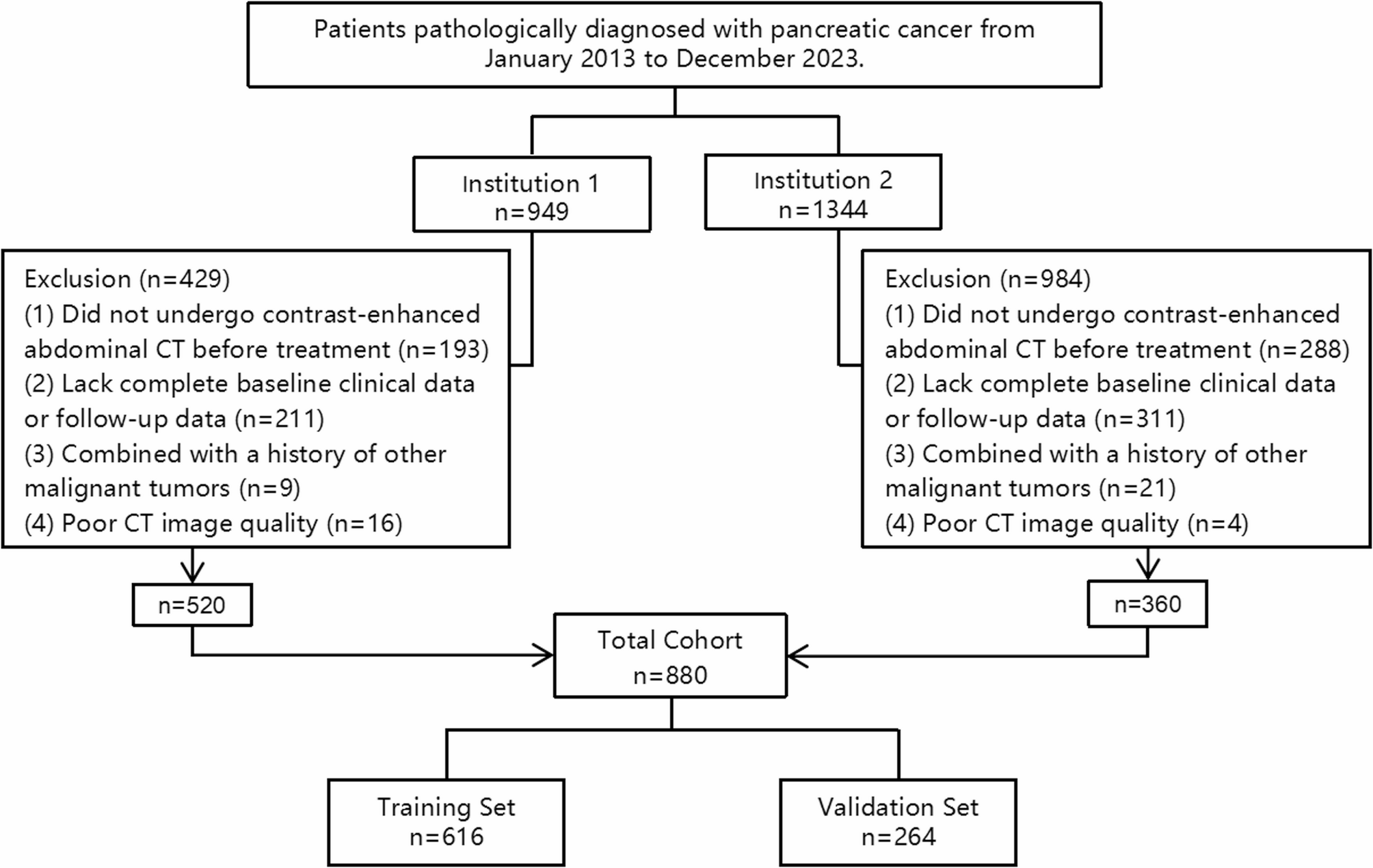
The study was approved by the Ethics Committee of the First Affiliated Hospital of Soochow University. A total of 880 patients diagnosed with pancreatic cancer between January 1, 2013, and December 31, 2023, were retrospectively collected from two hospitals, including 520 patients from the First Affiliated Hospital of Soochow University and 360 patients from the Second Affiliated Hospital of Soochow University. All patients received follow-up treatment in accordance with the CSCO guidelines for the diagnosis and treatment of pancreatic cancer [16]. The inclusion criteria were: (1) Histopathological diagnosis of pancreatic cancer; (2) Abdominal contrast-enhanced CT examinations performed at our hospital before definitive treatment. The exclusion criteria were: (1) Lack of complete baseline clinical data or follow-up data; (2) Combined with a history of other malignant tumors; (3) Poor quality of contrast-enhanced CT imaging, defined as scans with significant motion artifacts, insufficient contrast enhancement (i.e., poor visualization of pancreatic parenchyma and vessels during the portal venous phase), image blurring affecting tumor boundary visibility, or incomplete coverage of the pancreas within the scanning range. Patients were divided into a training set (n = 616) and a testing set (n = 264) in a 7:3 ratio. The specific selection process for the study cohort is shown in Fig. 1. Demographic characteristics such as gender and age were collected from electronic medical records. Tumor clinical staging was assessed according to the 8th edition of the AJCC (American Joint Committee on Cancer) staging system. Overall survival (OS) was followed up through telephone interviews and inpatient medical records, defined as the time from initial pathological diagnosis to death from any cause or the last known date of survival (Figure S 4). The follow-up was censored on June 1, 2024. OS was binarized as “survival” or “death” for machine learning classification. Death within 1-year, 2-year, and 3-year post-diagnosis was labeled as “death,” while survival at these time points was labeled as “survival.” Patients without follow-up information were already excluded according to the predefined exclusion criteria.
Fig. 1 Flowchart diagram shows the patient selection process from two medical centers. A total of 880 patients were included in this study and randomly divided into a training set (n = 616) and a test set (n = 264) at a ratio of 7:3. *Institution 1: The First Affiliated Hospital of Soochow University; Institution 2: The Second Affiliated Hospital of Soochow University
Imaging protocol and preprocessing
All patients underwent contrast-enhanced abdominal CT scans before initiating treatment. Imaging was performed using a 256-slice CT scanner, the GE Revolution CT from GE, USA, at the First Affiliated Hospital of Soochow University, and a 64-slice helical CT scanner, the GE Discovery CT 750 HD, at the Second Affiliated Hospital of Soochow University. Scan parameters were similar at both hospitals, with a tube voltage of 120 kV and a slice thickness and interslice gap of 5 mm. The Second Affiliated Hospital used automatic milliampere modulation, while the First Affiliated Hospital set the tube current between 200 and 450 mAs. Both hospitals used iodixanol 320 as the contrast agent, administered at a dose of approximately 1.4 mL per kilogram of body weight at an injection rate of 3 mL per second. Arterial phase images were acquired using an automatic triggering method, with a threshold of 100 Hounsfield units. Venous phase images were obtained after a 60-second delay.
Portal venous phase contrast-enhanced CT images in DICOM format were imported into 3D Slicer software (version 5.6.1). Guided by a senior radiotherapy oncologist (20 years of experience), a radiotherapy oncologist and a gastroenterologist (each with 3 years of experience) independently delineated tumor boundaries to create regions of interest (ROIs), carefully avoiding adjacent vessels, dilated bile ducts, and pancreatic ducts. Both physicians were blinded to the clinical outcomes prior to the ROI segmentation. If the two physicians produced significantly different ROIs for the same patient, a third radiologist (20 years of experience) reviewed the case and made the final decision in accordance with NCCN (National Comprehensive Cancer Network) guidelines. To assess the reliability and consistency of the ROI delineation, the intraclass correlation coefficient (ICC) was used for validation.
Radiomic feature extraction
Before feature extraction, the images were resampled using 3D Slicer software to a voxel size of 3 × 3 × 3 mm³ to ensure isotropy, thereby minimizing variability due to differences in scanning equipment and protocols. Radiomic features were then extracted using the Radiomics plugin within the 3D Slicer software. A total of 1,037 features were extracted, including 108 original features and 929 filtered features (LoG and wavelet), which were classified into seven categories: shape features, first-order (FO) features, features based on gray-level co-occurrence matrices (GLCM), gray-level dependence matrix (GLDM) features, features based on run-length matrices (RLM), features based on size-zone matrices (SZM), and features based on neighborhood gray-tone difference matrices (NGTDM).
The development and evaluation of models
Radiomics based model
To reduce the dimensionality of features and address the issue of multicollinearity, principal component analysis (PCA) was applied to the extracted radiomic features. We selected principal components that together explained 80% of the cumulative variance to minimize redundancy while retaining the most informative aspects of the original feature set. Subsequently, feature selection was further refined using the Least Absolute Shrinkage and Selection Operator (LASSO) regression, with the optimal λ value determined via 10-fold cross-validation. The final input features were determined by retaining principal components with non-zero coefficients.
The Random Survival Forest (RSF) algorithm, an ensemble tree-based method, is designed to analyze survival time data and predict individual survival probabilities or risks. RSF constructs models by integrating multiple survival trees, providing high stability and robustness while effectively reducing the risk of overfitting. Additionally, it captures complex non-linear relationships and automatically identifies key features closely associated with survival outcomes, thereby enhancing the interpretability of the model [17, 18]. In recent years, RSF has been widely applied to survival analysis problems [19]. In this study, the RSF algorithm was utilized to construct models in the training set for predicting 1-, 2-, and 3-year survival probabilities of patients. The RSF model was implemented using the randomForestSRC package in R. The number of trees (ntree) was set to 345, and the minimum terminal node size (nodesize) was optimized using the tune.nodesize() function, which selects the optimal value based on the out-of-bag (OOB) prediction error. The number of random splits (nsplit) was set to 11. The discriminatory ability of the models was evaluated using the receiver operating characteristic (ROC) curve and its area under the curve (AUC), while prediction consistency was assessed through accuracy measurements.
3D-Densenet based model
Given the importance of capturing three-dimensional structural information in medical imaging, we developed a deep learning model based on a three-dimensional densely connected convolutional network (3D-DenseNet) to predict 1-, 2-, and 3-year survival outcomes in pancreatic cancer patients. The 3D-DenseNet model was implemented using the MONAI (Medical Open Network for AI) framework, which is specifically designed for medical imaging analysis and provides robust deep learning tools optimized for healthcare applications.
The previously segmented tumor regions (ROI) were standardized to a fixed size of 96 × 96 × 96 voxels using linear interpolation to ensure uniform input dimensions and reduce computational complexity. To address the limitations of small sample datasets and enhance model robustness, image preprocessing was performed, including pixel intensity normalization, explicit channel dimension addition, and data type standardization. During the training phase, data augmentation techniques such as random 90-degree rotations, axis flipping, and Gaussian noise injection were applied to improve the model’s generalization capability.
The 3D-DenseNet architecture consisted of four densely connected blocks containing 6, 12, 24, and 16 convolutional layers, respectively. By leveraging dense connectivity, the model reused features from previous layers, improving feature learning efficiency and reducing the number of parameters. To extract both global and local features, a dual-input design (DualInputDenseNet) was adopted, integrating a whole-image branch and an ROI branch. The whole-image branch utilized 3D-DenseNet to extract global features, while the ROI branch employed two 3D convolutional layers, each followed by a ReLU activation function and max pooling operation, to progressively downsample and extract high-level ROI features. The outputs of both branches were concatenated along the channel dimension and passed through fully connected layers for feature fusion and final classification. The model was optimized using the Adam optimizer and trained for 100 epochs with a learning rate of 1 × 10⁻⁴. In each epoch, a batch loader was used to retrieve training data in batches. Cross-entropy loss was employed as the optimization objective, and model performance was monitored using test accuracy computed on the hold-out test set at each epoch. The model checkpoint that achieved the highest test set accuracy was saved for final evaluation. During the testing phase, data augmentation steps were disabled, and only normalization and resizing were applied to ensure stability and consistency in the evaluation process.
After training, the optimal 3D-DenseNet model was used to predict 1-, 2-, and 3-year survival probabilities for patients in both the training and test sets. AUC was calculated to assess the model’s discriminative ability. Additionally, accuracy was evaluated to measure classification consistency.
Fusion model
To further enhance the performance of the prognostic prediction model, this study built a fusion model by performing decision-level fusion of the radiomics model and the 3D-DenseNet model. The predictions from both models were combined with clinical features of patients (gender, age, T stage, N stage, and M stage) to predict the 1-, 2-, and 3-year survival outcomes. This approach aimed to fully leverage the different types of data to enhance the model’s predictive ability, thereby improving its generalizability and accuracy. To identify the optimal fusion model, four different machine learning algorithms were employed, including Logistic Regression, Random Forest, Support Vector Machine (SVM), and Decision Tree. The model performance was evaluated using accuracy and AUC.
After determining the optimal fusion model, patients were stratified based on mortality risk scores predicted by the model. Specifically, the optimal risk cutoff value was identified using X-tile software (version 3.6.1), and all patients were classified into low-risk and high-risk groups accordingly. Kaplan-Meier (KM) survival curves for the high-risk and low-risk groups were then plotted separately in both the training and test cohorts. A log-rank test was used to evaluate the statistical significance of survival differences between the two groups. Figure 2 provide an overview of the overall study workflow.
Fig. 2 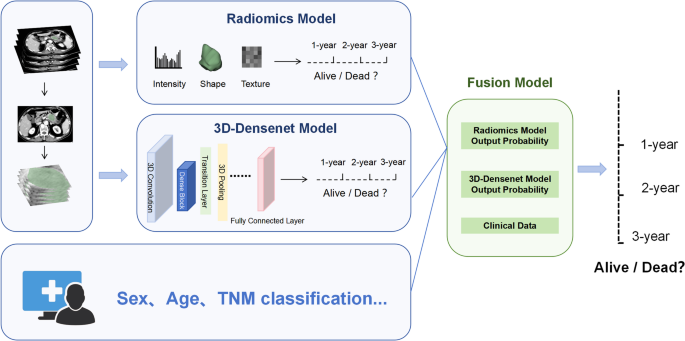
Workflow diagram for developing a predictive model. Tumor segmentation and region of interest (ROI) delineation were performed by two physicians. Quantitative features, including intensity, shape, and texture, were extracted from the ROI to train the radiomics model, while a deep learning model was trained based on the 3D-DenseNet framework. On this basis, a fusion model was constructed by integrating the predicted 1-year, 2-year, and 3-year survival probabilities from the radiomics model and 3D-DenseNet models along with baseline clinical characteristics of the patients. *3D-DenseNet, three-dimensional densely connected convolutional networks
Statistical analysis
All statistical analyses were performed using SPSS 26.0, R software (version 4.3.2), and Python 2.7.5. The demographic and clinical characteristics between the training and test sets were compared using SPSS 26.0. For continuous variables, an independent sample t-test was applied if the data followed a normal distribution, while the Mann-Whitney U test was used for non-normally distributed variables. Categorical variables were compared using the chi-square test. All statistical tests were two-sided, and a p-value < 0.05 was considered statistically significant. Feature selection, model construction, and evaluation of radiomics-based predictive models were performed using R (version 4.3.2). The “glmnet” package was employed for LASSO regression to select significant radiomic features. The “randomForestSRC” and “survival” packages were used to develop survival prediction models, while the “ggplot2” package was utilized for data visualization. The 3D deep learning model was constructed and evaluated using Python 2.7.5. The remaining general statistical analyses were conducted using R software.
Continue Reading
