Nico Hulkenberg conceded that Sauber’s strong show of pace at the United States Grand Prix was “unexpected”.
Hulkenberg was the surprise story in Sprint Qualifying, as he positioned his Sauber in fourth place. But he was unable to translate…

Nico Hulkenberg conceded that Sauber’s strong show of pace at the United States Grand Prix was “unexpected”.
Hulkenberg was the surprise story in Sprint Qualifying, as he positioned his Sauber in fourth place. But he was unable to translate…

Before every game at Emirates Stadium, Martin Odegaard lifts the lid on the mood in the camp via his captain’s notes published in the matchday programme.
Ahead of this evening’s big Champions League clash with Atletico Madrid, he discusses how…

SAN CLEMENTE, Calif., Oct. 21, 2025 /PRNewswire/ — Ease.io, a global leader in AI-powered software solutions that drive improvements in quality, safety, productivity, and compliance for manufacturers, today announced the…

Photo-Illustration: The Cut; Photos: Getty
When Ina Chung’s son was about 3 years old, he started…

Bayern began the 2025-26 campaign by winning the German Super Cup and they sit five points clear at the top of the Bundesliga.
“The contract extension with Vincent Kompany is a strong vote of confidence from the club after the great job he’s done…

giles AI, a company making advanced research tools accessible to all and simplifying how scientists work with data, has announced the commercial launch of giles, its AI specialised research agent designed to support healthcare,…
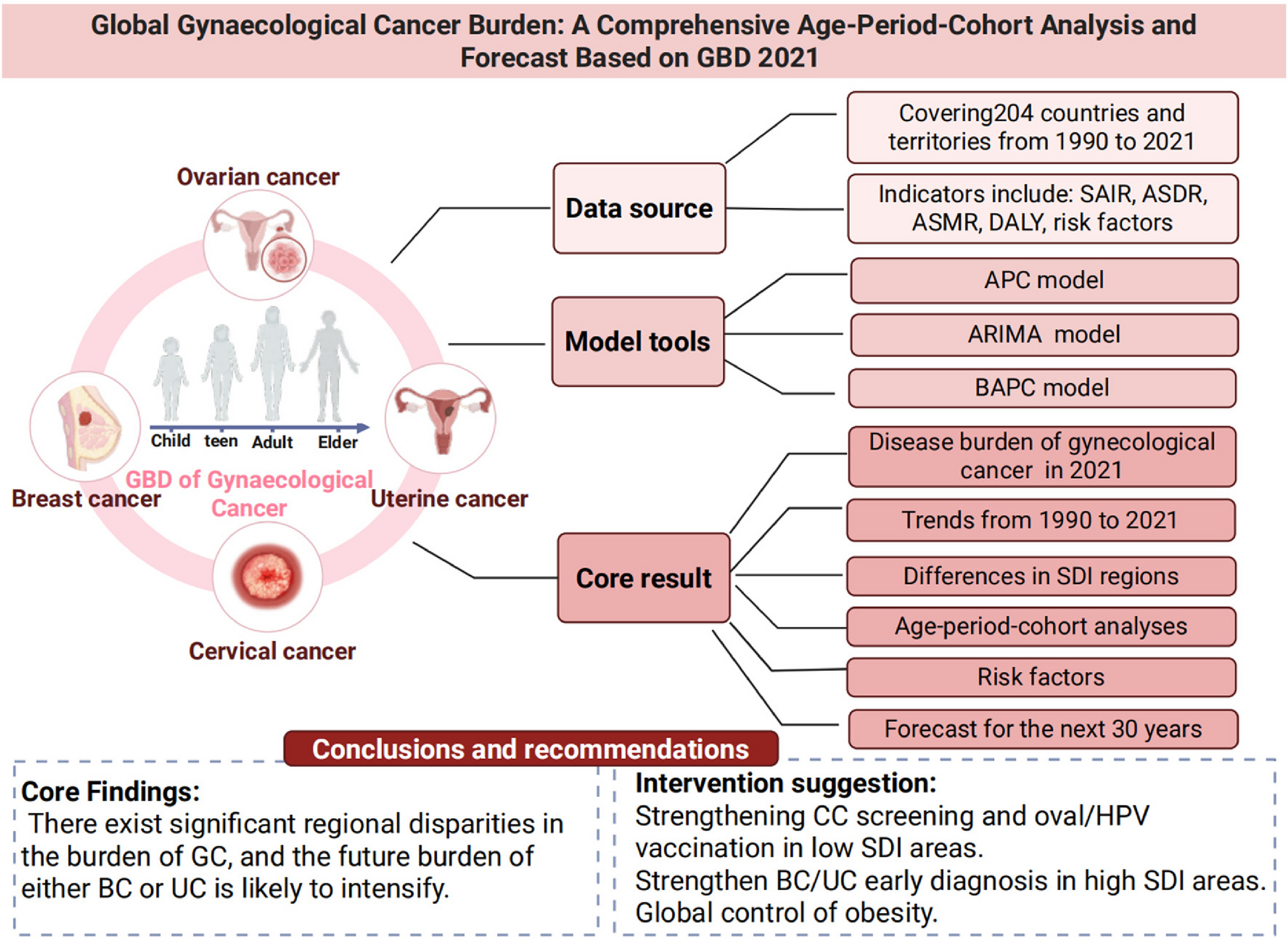
The global burden of GCs exhibits significant variation across cancer types, regions, and socio-economic contexts. Among these, BC remains the most prevalent and deadliest, with continued increases in…
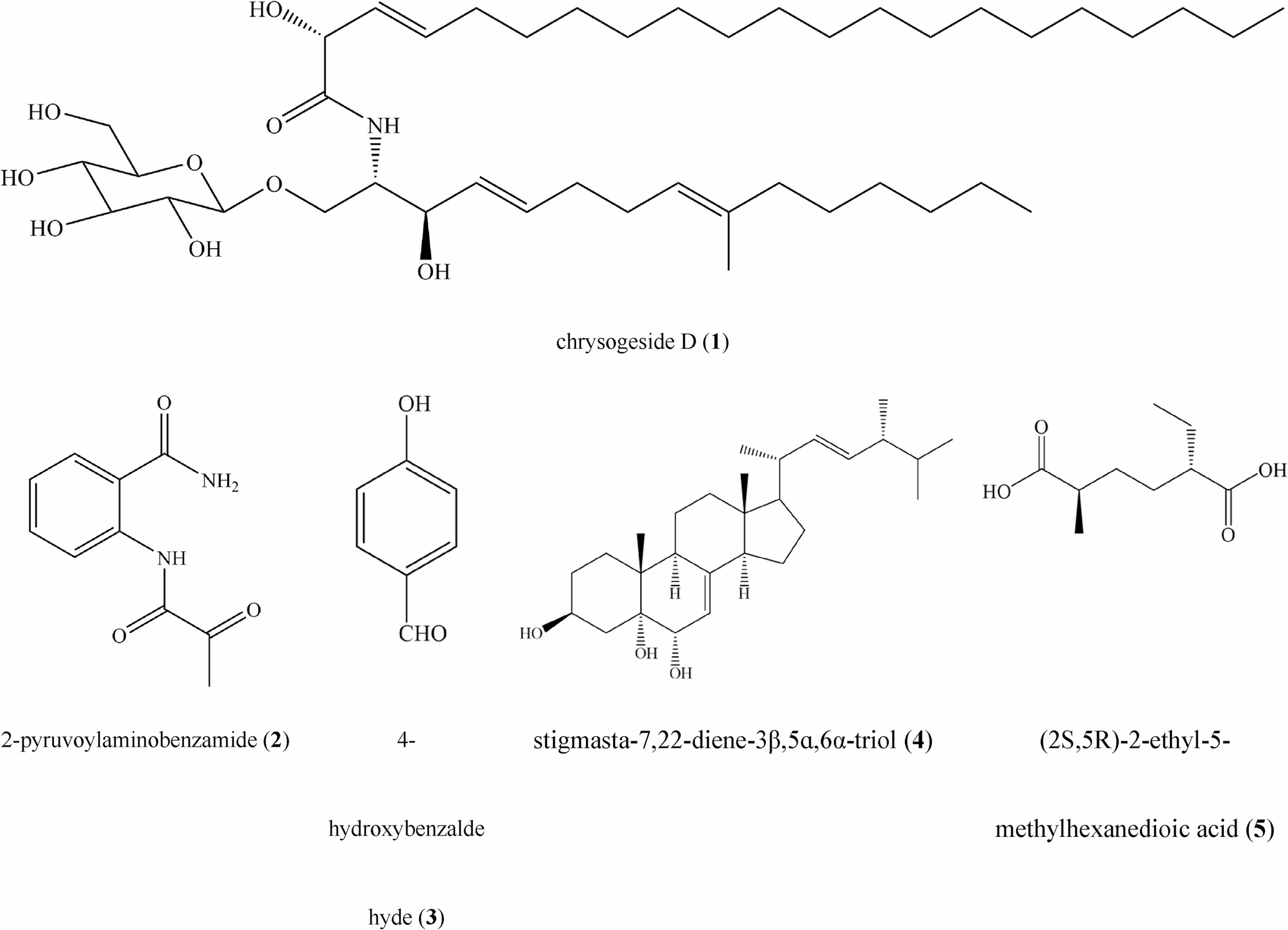
Beef extract and bacteriological peptone purchased from Guangdong Huankai Microbial Sci. & Tech. Co., Ltd., dextrose purchased from Xilong Science Co., Ltd., and agar powder purchased from Beijing Solarbio Science &…
Starzl TE. History of clinical transplantation. World J Surg. 2000;24(7):759–82.
Google Scholar
Cooper JE. Evaluation and treatment of acute rejection in kidney allografts. Clin J Am Soc Nephrol CJASN. 2020;15(3):430–8.
Google Scholar
Lusco MA, Fogo AB, Najafian B, Alpers CE. AJKD atlas of renal pathology: acute T-cell–mediated rejection. Am J Kidney Dis. 2016;67(5):e29-30.
Google Scholar
Naesens M, Roufosse C, Haas M, Lefaucheur C, Mannon RB, Adam BA, et al. The Banff 2022 kidney meeting report: reappraisal of microvascular inflammation and the role of biopsy-based transcript diagnostics. Am J Transplant. 2024;24(3):338–49.
Google Scholar
Loupy A, Haas M, Roufosse C, Naesens M, Adam B, Afrouzian M, et al. The Banff 2019 kidney meeting report (I): updates on and clarification of criteria for T cell– and antibody-mediated rejection. Am J Transplant. 2020;20(9):2318–31.
Google Scholar
Nankivell BJ, Agrawal N, Sharma A, Taverniti A, P’Ng CH, Shingde M, et al. The clinical and pathological significance of borderline T cell–mediated rejection. Am J Transplant. 2019;19(5):1452–63.
Google Scholar
Loupy A, Mengel M, Haas M. Thirty years of the International banff classification for allograft pathology: the past, present, and future of kidney transplant diagnostics. Kidney Int. 2022;101(4):678–91.
Google Scholar
de Freitas DG, Sellarés J, Mengel M, Chang J, Hidalgo LG, Famulski KS, et al. The nature of biopsies with “borderline rejection” and prospects for eliminating this category. Am J Transplant. 2012;12(1):191–201.
Google Scholar
Mehta RB, Tandukar S, Jorgensen D, Randhawa P, Sood P, Puttarajappa C, et al. Early subclinical tubulitis and interstitial inflammation in kidney transplantation have adverse clinical implications. Kidney Int. 2020;98(2):436–47.
Google Scholar
Heilman RL, Nijim S, Chakkera HA, Devarapalli Y, Moss AA, Mulligan DC, et al. Impact of acute rejection on kidney allograft outcomes in recipients on rapid steroid withdrawal. J Transplant. 2011;2011(1):583981.
Google Scholar
Masin-Spasovska J, Spasovski G, Dzikova S, Petrusevska G, Dimova B, Lekovski L, et al. The evolution of untreated borderline and subclinical rejections at first month kidney allograft biopsy in comparison with histological changes at 6 months protocol biopsies. Prilozi. 2005;26(1):25–33.
Google Scholar
Thierry A, Thervet E, Vuiblet V, Goujon JM, Machet MC, Noel LH, et al. Long-term impact of subclinical inflammation diagnosed by protocol biopsy one year after renal transplantation. Am J Transplant. 2011;11(10):2153–61.
Google Scholar
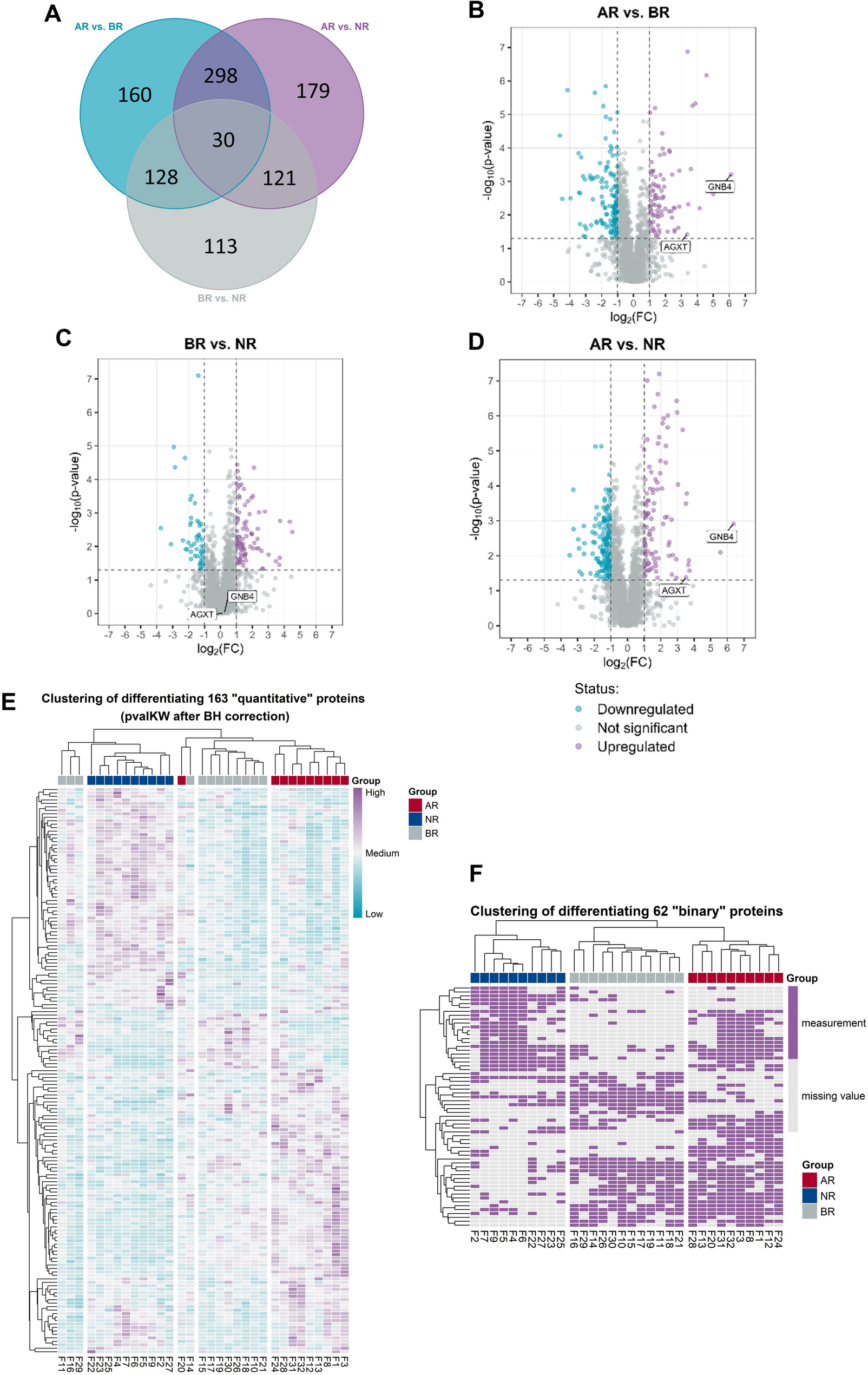
Wang C, Feng G, Zhao J, Xu Y, Li Y, Wang L, et al. Screening of novel biomarkers for acute kidney transplant rejection using DIA-MS based proteomics. PROTEOMICS – Clinical Appl. 2024;18(3):2300047.
Heidari SS, Nafar M, Kalantari S, Tavilani H, Karimi J, Foster L, et al. Urinary epidermal growth factor is a novel biomarker for early diagnosis of antibody mediated kidney allograft rejection: A urinary proteomics analysis. J Proteomics. 2021;30(240):104208.
Gwinner W, Karch A, Braesen JH, Khalifa AA, Metzger J, Naesens M, et al. Noninvasive diagnosis of acute rejection in renal transplant patients using mass spectrometric analysis of urine samples: a multicenter diagnostic phase III trial. Transplant Direct. 2022;8(5):e1316.
Google Scholar
Geoui T, Urlaub H, Plessmann U, Porschewski P. Extraction of proteins from formalin-fixed, paraffin-embedded tissue using the qproteome extraction technique and preparation of tryptic peptides for liquid chromatography/mass spectrometry analysis. Current Protocls Mol Biol. 2010;90(1):10–27.
Wiśniewski JR, Gaugaz FZ. Fast and sensitive total protein and Peptide assays for proteomic analysis. Anal Chem. 2015;87(8):4110–6.
Google Scholar
Conover WJ. Practical nonparametric statistics. 3rd ed. New York: Wiley; 1998.
Jonckheere AR. A distribution-free k-sample test against ordered alternatives. Biometrika. 1954;41(1–2):133–45.
Pallant J. SPSS Survival Manual: A Step by Step Guide to Data Analysis Using SPSS for Windows. 4th ed. McGraw Hill Open University Press; 2010.
Cohen J. Statistical Power Analysis for the Behavioral Sciences. 2nd ed. Hillsdale; 1988.
Armitage P. Tests for Linear Trends in Proportions and Frequencies. Biometrics. 1955;11(3):375.
Fang F, Liu P, Song L, Wagner P, Bartlett D, Ma L, et al. Diagnosis of T-cell-mediated kidney rejection by biopsy-based proteomic biomarkers and machine learning. Front Immunol. 2023;6(14):1090373.
Hofstraat R, Marx K, Blatnik R, Claessen N, Chojnacka A, Peters-Sengers H, et al. Highly Repeatable Tissue Proteomics for Kidney Transplant Pathology: Technical and Biological Validation of Protein Analysis using LC-MS/MS. bioRxiv; 2024. p 2024.06.14.599091.
Schmouder RL, Kunkel SL. The cytokine response in renal allograft rejection. Nephrol Dial Transplant. 1995;10(supp1):36–43.
Google Scholar
De Serres SA, Mfarrej BG, Grafals M, Riella LV, Magee CN, Yeung MY, et al. Derivation and validation of a cytokine-based assay to screen for acute rejection in renal transplant recipients. Clin J Am Soc Nephrol. 2012;7(6):1018.
Google Scholar
Karczewski J, Karczewski M, Glyda M, Wiktorowicz K. Role of TH1/TH2 cytokines in kidney allograft rejection. Transplant Proc. 2008;40(10):3390–2.
Google Scholar
Biro M, Munoz MA, Weninger W. Targeting Rho-GTPases in immune cell migration and inflammation. Br J Pharmacol. 2014;171(24):5491–506.
Google Scholar
Matsuda J, Asano-Matsuda K, Kitzler TM, Takano T. Rho GTPase regulatory proteins in podocytes. Kidney Int. 2021;99(2):336–45.
Google Scholar
Kurian SM, Velazquez E, Thompson R, Whisenant T, Rose S, Riley N, et al. Orthogonal comparison of molecular signatures of kidney transplants with subclinical and clinical acute rejection: equivalent performance is agnostic to both technology and platform. Am J Transplant. 2017;17(8):2103–16.
Google Scholar
Kurian SM, Williams AN, Gelbart T, Campbell D, Mondala TS, Head SR, et al. Molecular classifiers for acute kidney transplant rejection in peripheral blood by whole genome gene expression profiling. Am J Transplant. 2014;14(5):1164–72.
Google Scholar
Zhang M, Chen F, Feng S, Liu X, Wang Z, Shen N, et al. FBLN5 as one presumably prognostic gene potentially modulating tumor immune microenvironment for renal clear cell carcinoma in children and young adults. Pharmacogenomics Pers Med. 2024;19(17):27–40.
Huang W, Shi H, Hou Q, Mo Z, Xie X. GSTM1 and GSTT1 polymorphisms contribute to renal cell carcinoma risk: evidence from an updated meta-analysis. Sci Rep. 2015;5(1):17971.
Google Scholar
Li P, Li D, Lu Y, Pan S, Cheng F, Li S, et al. GSTT1/GSTM1 deficiency aggravated cisplatin-induced acute kidney injury via ROS-triggered ferroptosis. Front Immunol. 2024;25(15):1457230.
Beothe T, Docs J, Kovacs G, Peterfi L. Increased level of TXNIP and nuclear translocation of TXN is associated with end stage renal disease and development of multiplex renal tumours. BMC Nephrol. 2024;17(25):227.
Luczak M, Formanowicz D, Marczak Ł, Pawliczak E, Wanic-Kossowska M, Figlerowicz M, et al. Deeper insight into chronic kidney disease-related atherosclerosis: comparative proteomic studies of blood plasma using 2DE and mass spectrometry. J Transl Med. 2015;13(1):20.
Google Scholar
Abdullah EL, Jalalonmuhali M, Ng KP, Jamaluddin FA, Lim SK. The role of lymphocyte subset in predicting allograft rejections in kidney transplant recipients. Transplant Proceed. 2022;54(2):312–9.
Halloran PF. T cell-mediated rejection of kidney transplants: a personal viewpoint. Am J Transplant. 2010;10(5):1126–34.
Google Scholar
Liu B, Chen L, Huang H, Huang H, Jin H, Fu C. Prognostic and immunological value of GNB4 in gastric cancer by analyzing TCGA database. Dis Markers. 2022;16(2022):1–16.
Zhan J, Huang L, Niu L, Lu W, Sun C, Liu S, et al. Regulation of CD73 on NAD metabolism: unravelling the interplay between tumour immunity and tumour metabolism. Cell Commun Signal. 2024;22(1):387.
Google Scholar
Chen S, Wainwright DA, Wu JD, Wan Y, Matei DE, Zhang Y, et al. Cd73: an emerging checkpoint for cancer immunotherapy. Immunotherapy. 2019;11(11):983–97.
Google Scholar
Antonioli L, Pacher P, Vizi ES, Haskó G. CD39 and CD73 in immunity and inflammation. Trends Mol Med. 2013;19(6):355–67.
Google Scholar
Schneider E, Winzer R, Rissiek A, Ricklefs I, Meyer-Schwesinger C, Ricklefs FL, et al. CD73-mediated adenosine production by CD8 T cell-derived extracellular vesicles constitutes an intrinsic mechanism of immune suppression. Nat Commun. 2021;12(1):5911.
Google Scholar
Deutsch EW, Bandeira N, Perez-Riverol Y, Sharma V, Carver JJ, Mendoza L, et al. The proteomexchange consortium at 10 years: 2023 update. Nucleic Acids Res. 2023;51(D1):D1539–48.
Google Scholar
Perez-Riverol Y, Bandla C, Kundu DJ, Kamatchinathan S, Bai J, Hewapathirana S, et al. The PRIDE database at 20 years: 2025 update. Nucleic Acids Res. 2025;53(D1):D543–53.
Google Scholar