HIGH-RISK women had limited awareness yet strong support for breast cancer polygenic risk scores in clinical risk assessment more.
Study Snapshot
In a survey of 828 women at elevated breast cancer risk without a prior diagnosis, only 18.5% had…

HIGH-RISK women had limited awareness yet strong support for breast cancer polygenic risk scores in clinical risk assessment more.
In a survey of 828 women at elevated breast cancer risk without a prior diagnosis, only 18.5% had…
Chattaway MA, Gentle A, Nair S, Tingley L, Day M, Mohamed I, Jenkins C, Godbole G. Phylogenomics and antimicrobial resistance of Salmonella Typhi and paratyphi A, B and C in england, 2016–2019. Microb Genom 2021, 7(8):000633.
Xiang Y, Zhu K, Min K, Zhang Y, Liu J, Liu K, et al. Characterization of a Salmonella enterica serovar typhimurium lineage with rough colony morphology and multidrug resistance. Nat Commun. 2024. https://doi.org/10.1038/s41467-024-50331-y.
Google Scholar
Coburn B, Grassl GA, Finlay BB. Salmonella, the host and disease: a brief review. Immunol Cell Biol. 2007;85(2):112–8.
Google Scholar
Shi H, Zhou X, Zou W, Wang Y, Lei C, Xiang R, et al. Co-occurrence of biofilm formation and quinolone resistance in Salmonella enterica serotype typhimurium carrying an IncHI2-type oqxAB-positive plasmid. Microb Pathog. 2018;123:68–73.
Google Scholar
Chang MX, Zhang JF, Sun YH, Li RS, Lin XL, Yang L, et al. Contribution of different mechanisms to ciprofloxacin resistance in Salmonella spp. Front Microbiol. 2021;12:663731.
Google Scholar
Nambiar RB, Elbediwi M, Ed-Dra A, Wu B, Yue M. Epidemiology and antimicrobial resistance of Salmonella serovars typhimurium and 4,[5],12:i- recovered from hospitalized patients in China. Microbiol Res. 2024;282:127631.
Google Scholar
Kuang D, Zhang J, Xu X, Shi W, Chen S, Yang X, et al. Emerging high-level Ciprofloxacin resistance and molecular basis of resistance in Salmonella enterica from humans, food and animals. Int J Food Microbiol. 2018;280:1–9.
Google Scholar
Yu F, Chen Q, Yu X, Pan J, Li Q, Yang L, et al. High prevalence of plasmid-mediated quinolone resistance determinant aac(6’)-Ib-cr amongst Salmonella enterica serotype typhimurium isolates from hospitalised paediatric patients with diarrhoea in China. Int J Antimicrob Agents. 2011;37(2):152–5.
Google Scholar
Campillo R, Garcia-Penas I, Lopez N, Sanchez A, Fau A, Gomez D, et al. Ciprofloxacin-resistant Salmonella typhimurium demonstrates cross-tolerance to heat treatments in liquid food matrices. Food Res Int. 2025;210:116330.
Google Scholar
Andersson DI, Nicoloff H, Hjort K. Mechanisms and clinical relevance of bacterial heteroresistance. Nat Rev Microbiol. 2019;17(8):479–96.
Google Scholar
Lin JY, Zhu ZC, Zhu J, Chen L, Du H. Antibiotic heteroresistance in Klebsiella pneumoniae: definition, detection methods, mechanisms, and combination therapy. Microbiol Res. 2024;283:127701.
Google Scholar
El-Halfawy OM, Valvano MA. Antimicrobial heteroresistance: an emerging field in need of clarity. Clin Microbiol Rev. 2015;28(1):191–207.
Google Scholar
Chen Y, Hu D, Zhang Q, Liao XP, Liu YH, Sun J. Efflux pump overexpression contributes to Tigecycline heteroresistance in Salmonella enterica serovar typhimurium. Front Cell Infect Microbiol. 2017;7:37.
Google Scholar
Hjort K, Nicoloff H, Andersson DI. Unstable tandem gene amplification generates heteroresistance (variation in resistance within a population) to colistin in Salmonella enterica. Mol Microbiol. 2016;102(2):274–89.
Google Scholar
Weston N, Sharma P, Ricci V, Piddock LJV. Regulation of the AcrAB-TolC efflux pump in Enterobacteriaceae. Res Microbiol. 2018;169(7–8):425–31.
Google Scholar
Hernando-Amado S, Laborda P, La Rosa R, Molin S, Johansen HK, Martinez JL. Ciprofloxacin resistance rapidly declines in NfxB defective clinical strains of Pseudomonas aeruginosa. Nat Commun. 2025;16(1):4992.
Google Scholar
Suwanthada P, Kongsoi S, Jayaweera S, Akapelwa ML, Thapa J, Nakajima C, et al. Interplay between amino acid substitution in GyrA and QnrB19: elevating fluoroquinolone resistance in Salmonella typhimurium. ACS Infect Dis. 2024;10(8):2785–94.
Google Scholar
Strahilevitz J, Jacoby GA, Hooper DC, Robicsek A. Plasmid-mediated quinolone resistance: a multifaceted threat. Clin Microbiol Rev. 2009;22(4):664–89.
Google Scholar
Wong MH, Chan EW, Xie L, Li R, Chen S. IncHI2 plasmids are the key vectors responsible for OqxAB transmission among Salmonella species. Antimicrob Agents Chemother. 2016;60(11):6911–5.
Google Scholar
Wan Y, Myall AC, Boonyasiri A, Bolt F, Ledda A, Mookerjee S, Weisse AY, Getino M, Turton JF, Abbas H, et al. Integrated analysis of patient networks and plasmid genomes to investigate a regional, multispecies outbreak of Carbapenemase-Producing enterobacterales carrying both blaIMP and mcr-9 genes. J Infect Dis. 2024;230(1):e159–70.
Google Scholar
Treilles M, Chatre P, Drapeau A, Madec JY, Haenni M. Spread of the mcr-1 colistin-resistance gene in Escherichia coli through plasmid transmission and chromosomal transposition in French goats. Front Microbiol. 2022;13:1023403.
Google Scholar
Tan W, Lu Y, Zhu Z, Xu Z, Zhang Y, Huang Q, Meng X, Li S. Cotransfer of resistance to cephalosporins, colistin, and fosfomycin mediated by an IncHI2/pSH16G4928-like plasmid in ESBL-producing monophasic Salmonella typhimurium strains of pig origin. J Appl Microbiol 2023, 134(3):lxac060.
Deng L, Lv LC, Tu J, Yue C, Bai Y, He X, et al. Clonal spread of blaNDM-1-carrying Salmonella enterica serovar typhimurium clone ST34 and wide spread of IncHI2/ST3-blaNDM-5 plasmid in China. J Antimicrob Chemother. 2024;79(8):1900–9.
Google Scholar
Zhang CZ, Zhang Y, Ding XM, Lin XL, Lian XL, Trampari E, et al. Emergence of ciprofloxacin heteroresistance in foodborne Salmonella enterica serovar agona. J Antimicrob Chemother. 2020. https://doi.org/10.1093/jac/dkaa288.
Google Scholar
Zwe YH, Chin SF, Kohli GS, Aung KT, Yang L, Yuk HG. Whole genome sequencing (WGS) fails to detect antimicrobial resistance (AMR) from heteroresistant subpopulation of Salmonella enterica. Food Microbiol. 2020;91:103530.
Google Scholar
Langfelder P, Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics. 2008;9:559.
Google Scholar
Zhang B, Horvath S. A general framework for weighted gene co-expression network analysis. Stat Appl Genet Mol Biol. 2005;4:Article17.
Google Scholar
Zhang H, Ma S, Wang Y, Chen X, Li Y, Wang M, Xu Y. Development of an obesity-related multi-gene prognostic model incorporating clinical characteristics in luminal breast cancer. iScience. 2024;27(3):109133.
Google Scholar
Lieberman-Aiden E, van Berkum NL, Williams L, Imakaev M, Ragoczy T, Telling A, Amit I, Lajoie BR, Sabo PJ, Dorschner MO, et al. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science. 2009;326(5950):289–93.
Google Scholar
Lioy VS, Cournac A, Marbouty M, Duigou S, Mozziconacci J, Espeli O, et al. Multiscale structuring of the E. coli chromosome by nucleoid-associated and condensin proteins. Cell. 2018;172(4):771-e783718.
Google Scholar
Zhao X, Drlica K. Reactive oxygen species and the bacterial response to lethal stress. Curr Opin Microbiol. 2014;21:1–6.
Google Scholar
Hall CW, Zhang L, Mah TF. PA3225 is a transcriptional repressor of antibiotic resistance mechanisms in Pseudomonas aeruginosa. Antimicrob Agents Chemother. 2017. https://doi.org/10.1128/AAC.02114-16.
Google Scholar
Dewachter L, Fauvart M, Michiels J. Bacterial heterogeneity and antibiotic survival: understanding and combatting persistence and heteroresistance. Mol Cell. 2019;76(2):255–67.
Google Scholar
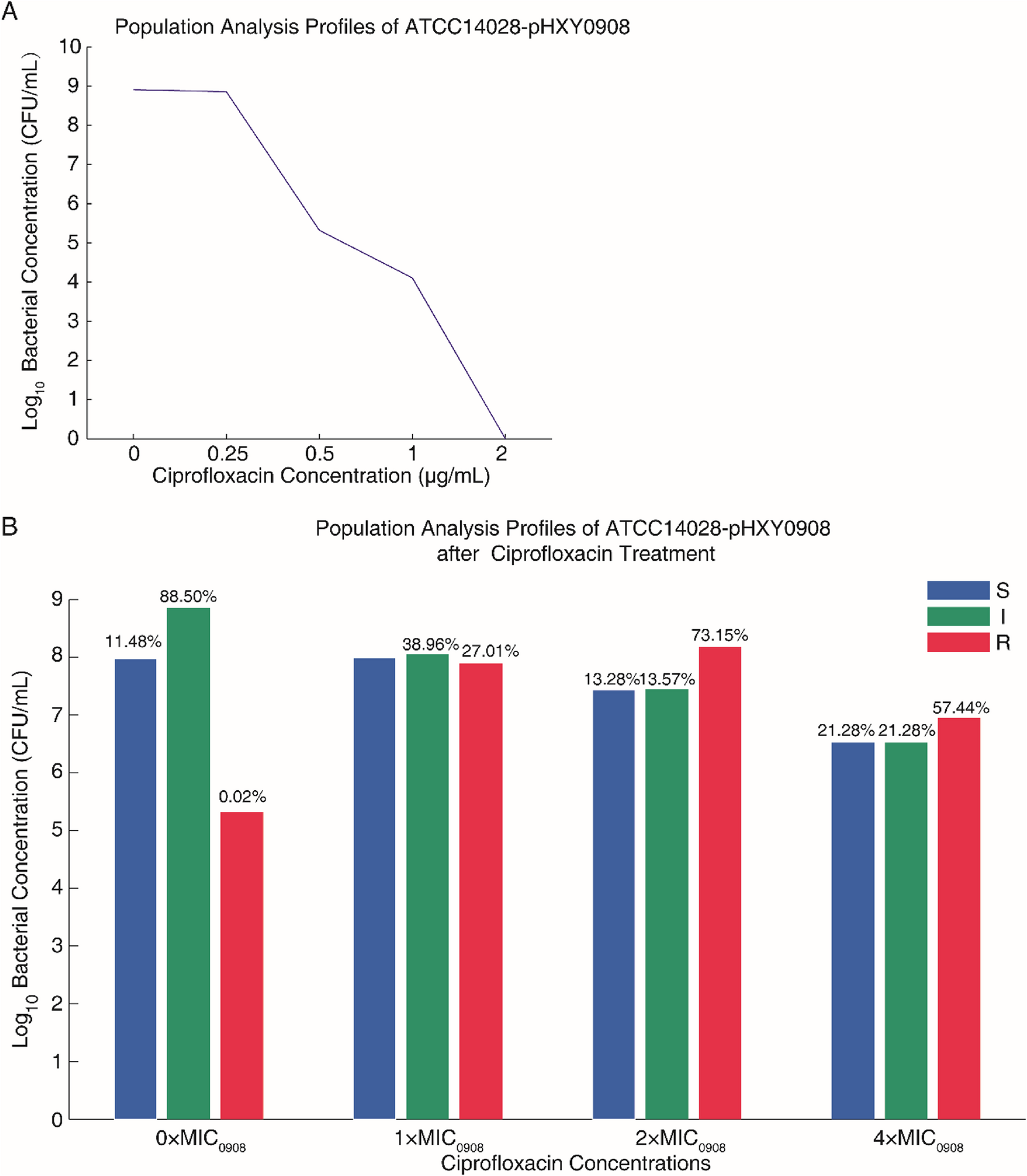
Lian X, Wang X, Liu X, Xia J, Fang L, Sun J, et al. oqxAB-positive IncHI2 plasmid pHXY0908 increase Salmonella enterica serotype typhimurium strains tolerance to ciprofloxacin. Front Cell Infect Microbiol. 2019;9:242.
Google Scholar
Calhoun LN, Kwon YM. Structure, function and regulation of the DNA-binding protein Dps and its role in acid and oxidative stress resistance in Escherichia coli: a review. J Appl Microbiol. 2011;110(2):375–86.
Google Scholar
Oh TJ, Jung IL, Kim IG. The Escherichia coli SOS gene SbmC is regulated by H-NS and RpoS during the SOS induction and stationary growth phase. Biochem Biophys Res Commun. 2001;288(4):1052–8.
Google Scholar
Labrou M, Michail G, Ntokou E, Pittaras TE, Pournaras S, Tsakris A. Activity of oxacillin versus that of vancomycin against oxacillin-susceptible mecA-positive Staphylococcus aureus clinical isolates evaluated by population analyses, time-kill assays, and a murine thigh infection model. Antimicrob Agents Chemother. 2012;56(6):3388–91.
Google Scholar
Wu X, Xu L, Gu W, Xu Q, He QY, Sun X, Zhang G. Iterative genome correction largely improves proteomic analysis of nonmodel organisms. J Proteome Res. 2014;13(6):2724–34.
Google Scholar
Zhang G, Fedyunin I, Kirchner S, Xiao C, Valleriani A, Ignatova Z. FansE: an accurate algorithm for quantitative mapping of large scale sequencing reads. Nucleic Acids Res. 2012;40(11):e83.
Google Scholar
Xiao CL, Mai ZB, Lian XL, Zhong JY, Jin JJ, He QY, et al. FANSe2: a robust and cost-efficient alignment tool for quantitative next-generation sequencing applications. PLoS ONE. 2014;9(4):e94250.
Google Scholar
Yang L, Lian X, Zhang W, Guo J, Wang Q, Li Y, Chen Y, Yin X, Yang P, Lan F, et al. Finding missing proteins from the epigenetically manipulated human cell with stringent quality criteria. J Proteome Res. 2015;14(9):3645–57.
Google Scholar
Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. Mapping and quantifying mammalian transcriptomes by RNA-seq. Nat Methods. 2008;5(7):621–8.
Google Scholar
Bloom JS, Khan Z, Kruglyak L, Singh M, Caudy AA. Measuring differential gene expression by short read sequencing: quantitative comparison to 2-channel gene expression microarrays. BMC Genomics. 2009;10:221.
Google Scholar
Zhao J, Zhang H, Qin B, Nikolay R, He QY, Spahn CMT, et al. Multifaceted stoichiometry control of bacterial operons revealed by deep proteome quantification. Front Genet. 2019;10:473.
Google Scholar
Robinson MD, McCarthy DJ, Smyth GK. EdgeR: a bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26(1):139–40.
Google Scholar
McCarthy DJ, Chen Y, Smyth GK. Differential expression analysis of multifactor RNA-seq experiments with respect to biological variation. Nucleic Acids Res. 2012;40(10):4288–97.
Google Scholar
Futschik ME, Carlisle B. Noise-robust soft clustering of gene expression time-course data. J Bioinform Comput Biol. 2005;3(4):965–88.
Google Scholar
Kumar L. Mfuzz: a software package for soft clustering of microarray data. Bioinformation. 2007;2(1):5–7.
Google Scholar
Ghazalpour A, Doss S, Zhang B, Wang S, Plaisier C, Castellanos R, Brozell A, Schadt EE, Drake TA, Lusis AJ, et al. Integrating genetic and network analysis to characterize genes related to mouse weight. PLoS Genet. 2006;2(8):e130.
Google Scholar
Mi H, Muruganujan A, Ebert D, Huang X, Thomas PD. PANTHER version 14: more genomes, a new PANTHER GO-slim and improvements in enrichment analysis tools. Nucleic Acids Res. 2019;47(D1):D419-26.
Google Scholar
Supek F, Bošnjak M, Škunca N, Šmuc T. Revigo summarizes and visualizes long lists of gene ontology terms. PLoS ONE. 2011;6(7):e21800.
Google Scholar
Ortet P, De Luca G, Whitworth DE, Barakat M. P2TF: a comprehensive resource for analysis of prokaryotic transcription factors. BMC Genomics. 2012;13:628.
Google Scholar

The Blue Sky thinking session at blooloop’s Festival of Innovation shared insights from guest speakers Kimberly Beneville, co-founder of Beneville Studios; Sam Bompas, director of Bompas & Parr; and Ben Wilson, head of architecture and concept…
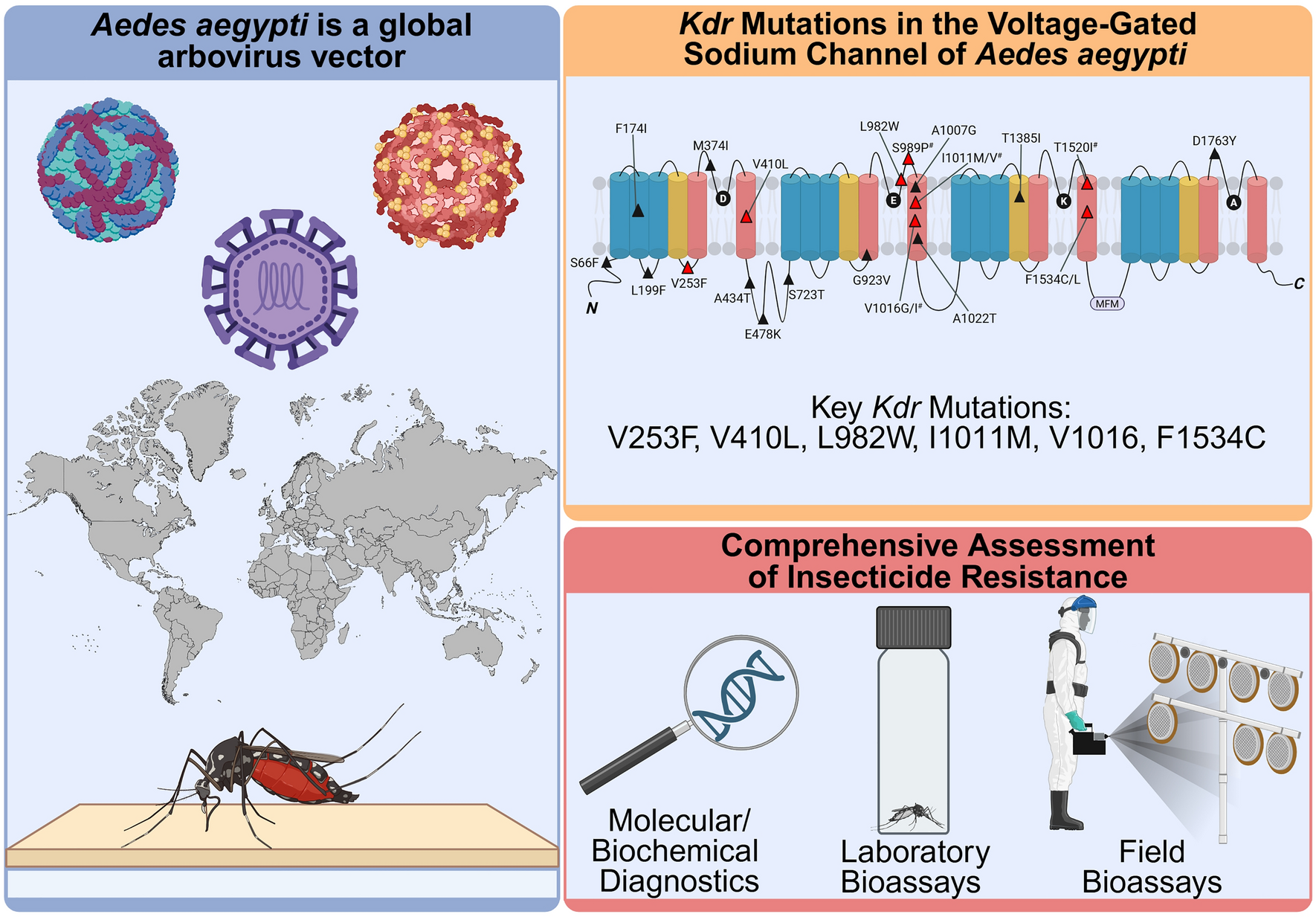
Messina JP, Brady OJ, Golding N, Kraemer MUG, Wint GRW, Ray SE, et al. The current and future global distribution and population at risk of dengue. Nat Microbiol. 2019;4:1508–15. https://doi.org/10.1038/s41564-019-0476-8.

Changes in eyesight are one of the most familiar effects of getting older. Sit in a dim restaurant with someone over 60, and you might hear, “Hold on — let me pull out my cell phone. I need more light to read the menu!” But what if declining…

Water quality is the most important thing for aquaculture farms to monitor to ensure their livestock remain healthy. While there are existing ways to monitor water quality — sensors and water testing kits — they are too expensive for many…
In a compelling Genomic Press Interview published today in Genomic Psychiatry, Dr. Bruce M. Cohen discusses results and insights that are reshaping international approaches to understanding and treating neuropsychiatric disorders….
This request seems a bit unusual, so we need to confirm that you’re human. Please press and hold the button until it turns completely green. Thank you for your cooperation!


Marvell ACC Linear Equalizers Enable Longer Reach and Power-efficient Copper in High-speed, Scale-up Interconnects
SANTA CLARA, Calif.–(BUSINESS WIRE)–
2025 OCP Global Summit — Marvell Technology, Inc. (NASDAQ: MRVL), a leader in data infrastructure semiconductor solutions, today announced that it is expanding its industry-leading connectivity portfolio with the addition of Marvell® active copper cable (ACC) linear equalizers.
The scale and complexity of today’s AI workloads are driving exponential growth in data center bandwidth, requiring new challenges in managing thermal and power efficiency. Copper remains the preferred solution for in-rack scale-up interconnects due to its low cost and ease of deployment. However, next-generation AI systems demand thinner copper-based interconnects within server racks to improve airflow and cooling. Meanwhile, as bandwidth and cable gauge requirements continue to rise, the signal transmission performance of direct attach copper (DAC) technology is increasingly limited.
Analog ACC devices incorporate a signal equalizer, offering longer reach than traditional passive DAC cables while adding minimal latency. They are also more cost-effective and power-efficient than digital alternatives.
Leveraging Marvell industry-leading PAM4 technology and expertise in 100G/lane and 200G/lane analog devices, the new Marvell ACC linear equalizers deliver superior gain, extending the reach of ACC compared to competitive ACC solutions at the same cable gauge. They support 800G and 1.6T copper interconnects and expand the Marvell scale-up interconnect portfolio, which includes chipsets for active electrical cables (AEC) and active optical cables (AOC).
“Offering a full complement of ACC, AEC and AOC silicon technologies, Marvell is unique in the scale-up interconnect landscape, providing customers with a full range of solutions to meet their individual requirements,” said Xi Wang, senior vice president and general manager, Connectivity Business Unit at Marvell. “We are excited to work with our ecosystem of cable OEM partners and system vendors to provide end customers with high-performance, in-rack connectivity solutions to handle their most advanced AI workloads.”
Marvell is showcasing its latest advancements in accelerated infrastructure at the OCP Global Summit this week, October 13 to 16, at the San Jose Convention Center in San Jose, California. More information about Marvell at OCP 2025 can be found here.
Availability
Marvell ACC linear equalizers are currently sampling to customers.
About Marvell
To deliver the data infrastructure technology that connects the world, we’re building solutions on the most powerful foundation: our partnerships with our customers. Trusted by the world’s leading technology companies for over 30 years, we move, store, process and secure the world’s data with semiconductor solutions designed for our customers’ current needs and future ambitions. Through a process of deep collaboration and transparency, we’re ultimately changing the way tomorrow’s enterprise, cloud and carrier architectures transform—for the better.
Marvell and the M logo are trademarks of Marvell or its affiliates. Please visit www.marvell.com for a complete list of Marvell trademarks. Other names and brands may be claimed as the property of others.
This press release contains forward-looking statements within the meaning of the federal securities laws that involve risks and uncertainties. Forward-looking statements include, without limitation, any statement that may predict, forecast, indicate or imply future events, results or achievements. Actual events, results or achievements may differ materially from those contemplated in this press release. Forward-looking statements are only predictions and are subject to risks, uncertainties and assumptions that are difficult to predict, including those described in the “Risk Factors” section of our Annual Reports on Form 10-K, Quarterly Reports on Form 10-Q and other documents filed by us from time to time with the SEC. Forward-looking statements speak only as of the date they are made. Readers are cautioned not to put undue reliance on forward-looking statements, and no person assumes any obligation to update or revise any such forward-looking statements, whether as a result of new information, future events or otherwise.
View source version on businesswire.com: https://www.businesswire.com/news/home/20251014066478/en/
Media Contact:
George Millington
pr@marvell.com
Source: Marvell Technology, Inc.
Released October 14, 2025