Establishment of T2DM rats fracture model and rats treatments
The work has been reported in line with the ARRIVE guidelines 2.0.
After adapting to feeding for three days, the Sprague Dawley (SD) rats were weighed and classified by cage (3 rats/cage), and 60 male SD rats of 3 weeks old were randomly distributed into 2 groups. Many studies have shown the procedures of establishing high-fat fed (HFD) and streptozotocin (STZ)-induced diabetic rat model which are a model of T2DM. According to the procedures of induction of the T2DM rat model in the existing study, after 3 weeks of feeding HFD (containing 60% fat), rats in the T2DM group were injected with STZ (40 mg/kg in citrate buffer). The control group fed with a normal pelleted diet received an equal volume of citrate buffer [21, 22].
The blood glucose (BG) levels were evaluated consecutively, and blood samples were collected from the tail vein. According to the procedures, rats with randomly blood glucose (RBG) samples > 16.7 mmol/L more than three times were identified to have T2DM after 7 weeks. During diabetes induction, animals were given free access to their original diets (received the high-fat or control diet and water ad libitum) for 12 weeks.
To assess the T2DM model, we measured the metabolic index, including body weight, food intake, water consumption and volume of excreted urine at several time points, including before being fed the HFD and 12 weeks after STZ injection. In addition, RBG was observed at several special time points, which included before fed HFD, before STZ injection, one week after STZ injection, 6 weeks after STZ injection and 12 weeks after STZ injection. At the end of the observation, IPGTT and ITT were evaluated. In detail, to execute IPGTT, animals were fasted for 12 h and injected with 1.5 g/kg glucose. BG was measured at 0, 30, 60 and 120 min after glucose injection. While ITT was carried out by injecting the rats with 0.75 IU/kg insulin, and then BG was obtained at 0, 30, 60 and 120 min after insulin administration. Animals with an RBG below 10 mmol/L at any time points were regarded as nondiabetic, and those with an RBG between 10 and 16.7 mmol/L were excluded. At 12 weeks after STZ injection, rats remaining in the T2DM group were evaluated as diabetic rats in a model of T2DM [23, 24].
After 15 weeks of treatment, all the successfully induced T2DM rats received general anesthesia with pentobarbital sodium intraperitoneal injection before surgery. A lateral incision was made along the proximal femur, followed by longitudinal dissection of the skin, subcutaneous tissue, and muscle along the femoral axis. The surrounding soft tissues were gently separated to expose the femur. A transverse osteotomy of the mid-diaphysis of the femur was performed by an oscillating mini-saw to establish a transverse femur shaft fracture model. The knee was bent 90 degrees, the lateral patellar incision was made, and the Kirschner needle was inserted femur intramedullary through the femoral intercondylar groove. The tip of the Kirschner needle was inserted through the top of the femoral greater trochanter to stabilize the fracture. Finally, all the incisions were closed using a 4-0 nylon suture. All the rats were kept in individual cages. Unprotected weight bearing was allowed immediately after the operation. After surgery, all the animals were given food and water ad libitum. At the end of the experiments, a sodium pentobarbital solution was administered intraperitoneally at a dose of 200 mg/kg. Following administration, animals were carefully monitored until the complete cessation of respiration and heartbeat, and a secondary physical method was applied when necessary to confirm death. All procedures were performed by personnel trained in rodent euthanasia.
Based on X-ray examinations, the fracture sites in the rats were located and marked on their skin. Then, 600 µL of ADSC-exosomes induced by mechanical stretch at a concentration of 200 µg/mL, as well as an equal volume of PBS, were locally injected into the fracture site every three days after surgery, with 200 µL administered each time. Finally, X-ray examination, micro-CT examination, histochemistry analysis, and Western blot analysis of the fractured femurs were performed 28 days after the operation.
Cells obtaining and culture
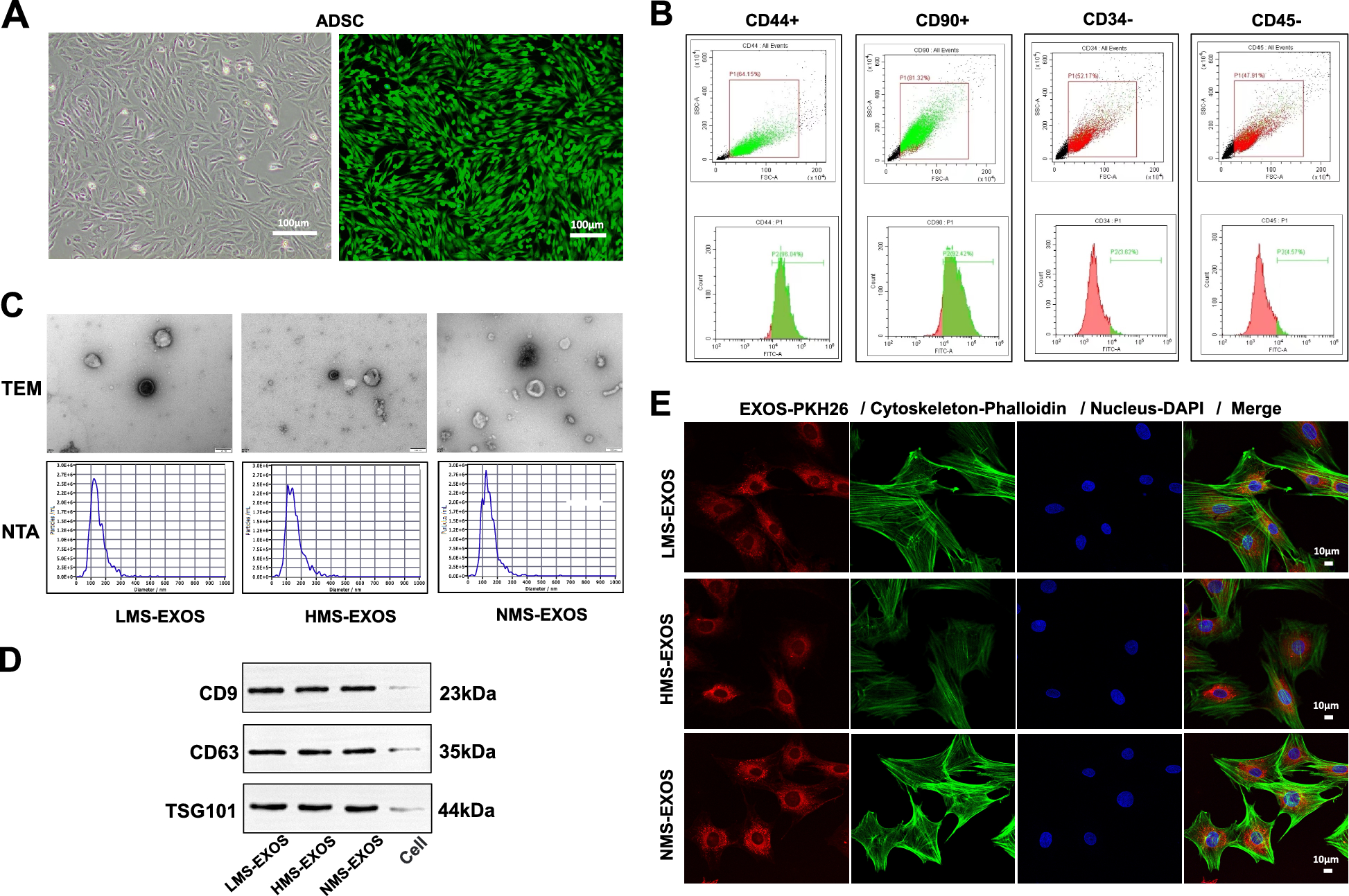
To obtain ADSCs, the subcutaneous fat in the groin of 3 weeks old male rats was cut into pieces as small as possible and centrifuged at 1500 rpm for 10 min to extract the sediment and glue enzyme was added in to digest at 37 °C for 40 min, then added the medium to terminate the reaction, and the mixture was filtered with a 70 μm filter. After centrifugating at 1500 rpm for 8 min, the cells were suspended in the medium and cultured in vitro for about 8 days. ADSCs (CD44+ CD90+ CD34-CD45-) were collected and certified by flow cytometry and the activity of ADSCs was determined by Calcein Acetoxymethyl Ester (Calcein AM) (Beyotime Biotechnology, Shanghai, China) staining assay.
Bone marrow cells were extracted from the femur and tibia of 3-week-old male rats, then isolated and cultured for 3 generations. BMSCs (CD44+ , CD34-) were identified by flow cytometry, and the cellular bioactivity of BMSCs was determined by Calcein AM staining assay.
The HUVECs (PUMC-HUVEC-T1; Cat NO: CL-0675) used in our experiments were originally purchased from a commercial vendor (Wuhan Pricella Biotechnology Co., Ltd.). To mimic the true condition and environment of Type 2 Diabetes Mellitus. All cells used in cellular function experiments were cultured with a complete culture medium consisting of high-concentration glucose (30 mM).
Tensile strain treatments of cells
Cell’s mechanical stimulation device has become a well-established method to apply mechanical strain to cultured cells; numerous similar devices for cell stimulation have been developed. In this study, external tensile strain was applied to ADSCs using a cell tension culture system. The special elastic membrane or well of the cell tension culture system was precoated with Matrigel (Corning, Bedford, MA, USA), incubated overnight, and kept moist with PBS for later use. ADSCs were cultured on the coated membrane or well. Cyclic tensile strain was applied using a uniaxial elongation model either 24 h post-seeding or upon reaching 90–100% confluence to simulate mechanical stimulation. In the selected uniaxial elongation model, cyclic sinusoidal tensile strain at a fixed frequency was applied to the cells according to a predefined protocol. All the ADSCs were divided into 3 groups, in which ADSCs were given of 1.0 Hz 6% (lower magnitude mechanical stretch ADSCs, LMS), 1.0 Hz 18% (higher magnitude mechanical stretch ADSCs, HMS) tensile strain, 2 h per day separately, and a total of 3 days. In the non-stress group (non-mechanical stretch ADSCs, NMS), 0% tensile strain was applied to ADSCs for 3 days in succession [25,26,27].
Isolation and identification of ADSC exosomes
After exposure to the corresponding tensile strain, ADSCs were washed three times with PBS and then cultured in exosome-free, FBS-free basal medium for an additional 24 h. Exosomes were isolated using the technique of differential centrifugation. In detail, the supernatant was collected and centrifuged at 1000×g at 4 °C for 10 min, 2000×g at 4 °C for 10 min, then centrifuged at 10000×g at 4 °C for 30 min, and centrifuged at 140000×g at 4 °C for 90 min in turn using a Beckman Coulter ultracentrifuge (Beckman Coulter, USA). Discard supernatant PBS washing sample, 140000×g centrifuge for 90 min at 4 °C. The precipitated exosomes were collected and re-suspended in 0.5 mL PBS and conserved at − 80 °C [28]. Then the volume of exosomes was concentrated to 200 μL. The morphology of exosomes was observed by transmission electron microscope (TEM; HITACHI, HT7700, JAPAN) and the diameter distribution was analyzed by Nanoparticles Tracking Analysis (NTA; ZetaView, Particle Metrix, Meerbusch, Germany). Western blot analysis identified its specific biomarkers (CD9, CD63, TSG101).
Flow cytometry (FCM) assay
ADSCs were analyzed by flow cytometry. ADSCs were detected with specific biomarker antibodies CD44 (Affinity), CD90 (Abcam), CD34, and CD45 (Abcam). BMSCs were certified by specific biomarkers CD44 (Affinity) and CD34 (Abcam). Results were analyzed using Flowjo software (version 10.0; BD Biosciences).
Exosome uptake assay
Based mainly on the manufacturer’s standard procedure, PKH26 was used as a dye to label the exosomes in the exosome uptake assay. In short, exosomes were obtained with differential centrifugation (140,000×g, 20 min, 4 °C) dyed with PKH26 (mixed solution was incubated for 20 min at room temperature). While BMSCs were seeded onto a 35 mm confocal dish at the proper density and labeled with DAPI dye. Then, exosomes labeled with PKH26 were mixed and co-cultured with BMSCs labeled with DAPI for 12 h and finally observed via a confocal laser scanning microscopy (CLSM, Leica Microsystems, Germany).
ALP activity and mineralization assessment of osteogenic differentiation
Following three passages of culture, BMSCs were seeded into 6-well plates (2 × 105 cells per well) that had been pre-coated with a 0.1% gelatin solution. The plates were then incubated for 14 days using a specific osteogenic induction medium (Cyagen Biosciences). To evaluate the effect of ADSCs-Exos on osteogenic differentiation, 200 µL of ADSCs-Exos (LMS-Exos, NMS-Exos, HMS-Exos derived from ADSCs) with a concentration of 200 µg/mL and equal volumes of PBS were added into every well severally with the osteogenic induction medium, and the medium was refreshed every three days. To evaluate the level of osteogenic differentiation, the cells were stained with alizarin red staining (ARS) dye and alkaline phosphatase (ALP) staining, and were collected for Western Blotting analysis on day 14. In detail, BMSCs were osteogenic induced for 14 days with different treatments, then cells were washed two times via PBS, then fixed with 4% paraformaldehyde for 30 min at room temperature prepare for ALP and ARS staining. A BCIP/NBT ALP kit (Beyotime, China) was used for ALP staining. After the stained cells were washed using PBS three times, the BCIP/NBT substrate was utilized to stain osteogenic-induced BMSCs. The results were observed and imaged via optical microscopy and then calculated and evaluated by the Image J software processing system (NIH, USA), and GraphPad Prism 10.0 (GraphPad Software, CA, USA) was used for the data analysis.
Tube formation assay of ADSC-Exos
HUVECs were cultured in Matrigel (Corning, USA) precoated 24-well plates (1 × 105 cells per well). 200 µL of ADSCs-Exos (LMS-Exos, NMS-Exos, HMS-Exos derived from ADSCs) with a concentration of 200 µg/mL and equal volumes of PBS were added into every well with the medium, respectively. 6 h later, tube formation was observed with a fluorescence microscope. The Image J software processing system (NIH, USA) and GraphPad Prism 10.0 (GraphPad Software, CA, USA) were used for quantification and data analysis.
Scratch test and migration test
HUVECs were cultured in 6-well plates (2 × 105 cells per well). 200 µL of ADSCs-Exos (LMS-Exos, NMS-Exos, HMS-Exos derived from ADSCs) with a concentration of 200 µg/mL and equal volumes of PBS were added into every well with the medium without serum. 24 h later when cultured HUVEC cells reached 100% confluence, a straight scratch was made with a 200 µL pipette in every well. 0 h, scratched wounds were observed with a fluorescence microscope and recorded. 12 h later, Scratch wound healing results were observed with a fluorescence microscope and photos were taken. Before observation, all samples were stained with calcein AM dye (Beyotime Biotechnology, Shanghai, China). The Image J software processing system (NIH, USA) and GraphPad Prism 10.0 (GraphPad Software, CA, USA) were used for quantification and data analysis.
Transwell assay
HUVECs treated with 200 µL of ADSCs-Exos (LMS-Exos, NMS-Exos, HMS-Exos derived from ADSCs) with a concentration of 200 µg/mL and equal volumes of PBS were seeded into 24-well transwell of 8 µm pore diameter cell culture plate (Corning, USA). 12 h later, HUVECs were stained with crystal violet for 20 min, and then observed by an optical microscope. The Image J software processing system (NIH, USA) and GraphPad Prism 10.0 (GraphPad Software, CA, USA) were used for quantification and the data analysis.
Cell alive/dead assays
All cells, including ADSCs, HUVECs, and BMSCs, were stained before use via calcein AM and Propidium iodide (PI) dye (Beyotime Biotechnology, Shanghai, China) to evaluate cell vitality.
Western blot analysis
Western blotting was performed following previously described protocols. First, after the concentrations of protein were measured via BCA (Aspen), the protein was separated into equal amounts via SDS-PAGE (Beyotime, China), transferred into the PVDF membrane (Millipore, Burlington, MA, USA) and then incubated with 5% bovine serum albumin for 1 h at 25 °C. Next, the membranes were incubated overnight with primary antibodies specific for CD9 (Abcam), CD63 (Abcam), TSG101 (Abcam), Runx2 (Abcam), OCN (Santa), and GAPDH (Abcam). HRP-labeled secondary antibody (Abcam, USA) was added, and then, the membrane was visualized using a T5200 Multi Chemiluminescence Detection System (Tanon, China) as recommended of the manufacturer. The membranes were incubated with Immobilon ECL reagent (Thermo Fisher Scientific, Waltham, MA, USA), and the bands were quantified via Image J software (NIH, USA). GAPDH protein level was used as an internal control for MACF1. The Image J software processing system (NIH, USA) and GraphPad Prism 10.0 (GraphPad Software, CA, USA) were used for quantification of protein level and the data analysis. The significance level was set to a 95% confidence interval, and statistical significance was declared as * p < 0.05, ** p < 0.01, *** p < 0.001, **** p < 0.0001 and ns p > 0.05.
Micro-CT analysis
The animals were euthanized 4 weeks postoperatively, the internal fixations were removed and their femurs were fixed in 4% paraformaldehyde for 24 h at 4 °C. Then, tissue specimens were scanned via a micro-CT system (SkyScan1276, Bruker, Belgium) at a resolution of 9.054604 μm with 85 kV and 200 µA. After scanning, 3D structures of femurs were performed (Reconstruction was accomplished by NRecon (version 1.7.4.2)), and the new bone volume/total volume (BV/TV) were calculated to assess bone regeneration in the fracture site to assess the morphology of the reconstructed femurs (3D and 2D analysis were performed using software CT Analyser (version 1.20.3.0)).
qRT‑PCR analysis
Total RNA was isolated from exosomes using TRIZOL Extraction Reagent (G3013, Servicebio). The cDNA was reverse transcribed using RevertAid First Strand cDNA Synthesis Kit (Invitrogen, CA, USA) following the manufacturer’s instructions. And the qRT-PCR for mRNAs was performed on a StepOne™ Real-Time PCR System (Life technologies) using EnTurboSYBR Green PCR SuperMix (EQ001, ELK Biotechnology) or HieffTM qPCR SYBR™ Green Master Mix (No Rox Plus) (11201ES, Shanghai Yeasen BioTechnologies). The relative expression levels of mRNA or miRNA were normalized to those of GAPDH or U6 and evaluated using the 2−ΔΔCT method. The primers used for qRT-PCR are listed in Table 1.
Transfection
Following manufacturer’s protocols, miR-877 mimics or inhibitors and their NCs (Generalbiol, HeFei, China) were transfected into BMSCs and HUVECs to evaluate miR-877 function. After 48 h of transfection, the expression level of miR-877 was measured by qRT-PCR.
RNA sequencing and bioinformatics analysis
All the samples were processed as description previously. All the experiment procedures were according manufacture’s protocols and recommendations. Briefly, Total RNA was extracted from exosomes using TRIzol reagent (Invitrogen, CA, USA) according to the manufacturer’s protocol. Purity, concentration and integrity of RNA sample were examined by NanoDrop, Qubit 2.0, Agilent 2100, etc. RNA concentration and purity was measured using NanoDrop 2000 (Thermo Fisher Scientific, Wilmington, DE). RNA integrity was assessed using the RNA Nano 6000 Assay Kit of the Agilent Bioanalyzer 2100 system (Agilent Technologies, CA, USA). Only RNA with good quality could move on to following procedures. Total RNA from each sample was used to prepare miRNA library using NEB Next Ultra small RNA Sample Library Prep Kit (NEB, Boston, MA, USA) according to the Illumina small RNA sample preparation protocol. Sequencing was then performed on the Illumina novaseq6000 platform (Illumina, San Diego, CA). A total amount of 1.5 μg RNA per sample was used as input material for the RNA sample preparations. Briefly, first of all, ligated the 3′SR Adaptor. Mixed 3′SR Adaptor for Illumina, RNA and Nuclease-Free Water, after mixture system incubation for 2 min at 70 °C in a preheated thermal cycler, the Tube was transfer to ice. Then, add 3′ Ligation Reaction Buffer (2X) and 3′Ligation Enzyme Mix ligate the 3′SR Adaptor. Incubated for 1 h at 25 °C in a thermal cycler. To prevent adaptor-dimer formation, the SR RT Primer hybridizes to the excess of 3′SR Adaptor (that remains free after the 3′ligation reaction) and transforms the single stranded DNA adaptor into a double-stranded DNA molecule. And dsDNAs are not substrates for ligation mediated. The second, ligated the 5′SR Adaptor. Then, reverse transcription synthetic first chain. Last, PCR amplification and Size Selection. PAGE gel was used for electrophoresis fragment screening purposes, rubber cutting recycling as the pieces to get small RNA libraries. At last, PCR products were purified (AMPure XP system) and library quality was assessed on the Agilent Bioanalyzer 2100 system. The clustering of the index-coded samples was performed on a cBot Cluster Generation System using TruSeq PE Cluster Kit v4-cBot-HS (Illumia) according to the manufacturer’s instructions. After cluster generation, the library preparations were sequenced on an Illumina Hiseq 2500 platform and paired-end reads were generated. Raw data (raw reads) of fastq format were firstly processed through in-house perl scripts. In this step, clean data (clean reads) were obtained by removing reads containing adapter, ploy-N and low-quality reads from raw data. And reads were trimmed and cleaned by removing the sequences smaller than 18 nt or longer than 30 nt. Differential expression analysis of two groups was performed using the DESeq R package (1.10.1). DESeq provide statistical routines for determining differential expression in digital miRNA expression data using a model based on the negative binomial distribution. The resulting P values were adjusted using the Benjamini and Hochberg’s approach for controlling the false discovery rate. miRNA with an adjusted p < 0.05 found by DESeq were assigned as differentially expressed. Target gene function was annotated based on the following databases:Nr (NCBI non-redundant protein sequences); Nt (NCBI non-redundant nucleotide sequences); Pfam (Protein family); KOG/COG (Clusters of Orthologous Groups of proteins); Swiss-Prot (A manually annotated and reviewed protein sequence database); KO (KEGG Ortholog database); GO (Gene Ontology).
Histological and immunofluorescence analysis
The collected femurs of rats from different groups were fixed in 4% paraformaldehyde solution for 48 h, decalcified with 20% EDTA at 25 °C for 28 days, embedded in paraffin, and sectioned along the longitudinal axis. Sections from the fracture region were stained with hematoxylin and eosin (H&E), safranin O-fast green, Masson, RUNX2, OCN for immunohistochemical analysis. Immunofluorescence staining with α-SMA, CD31, RUNX2, OCN for immunofluorescence analysis. The sections were imaged and observed by a microscope.
Statistical analysis
All the experiments were performed at least three replicates per group. Values were presented as mean ± SD, and were analyzed with GraphPad Prism 10.0 (Graph-Pad Software, CA, USA). Variances between groups were assessed by the two-sided Student’s t-test (for two-group comparisons) or the one-way analysis of variance (ANOVA) with Tukey’s test (for more than two-group comparisons). And statistical significance was declared as* p < 0.05; ** p < 0.01; *** p < 0.001; **** p < 0.0001; ns p > 0.05, not significant.