Ireland will be without lock Joe McCarthy and wing Mack Hansen because of foot injuries, while centres Robbie Henshaw (groin) and Bundee Aki (hip) have emerged as doubts for the New Zealand game.
However, Fogarty remains hopeful that captain…

Ireland will be without lock Joe McCarthy and wing Mack Hansen because of foot injuries, while centres Robbie Henshaw (groin) and Bundee Aki (hip) have emerged as doubts for the New Zealand game.
However, Fogarty remains hopeful that captain…


Mosquitoes have been found in Iceland for the first time as global heating makes the country more hospitable for insects.
The country was until this month one of the only places in the world that did not have a mosquito population. The other is…

Read more news stories here
Dr Louis van der Watt, a well-respected string teacher in the South African classical music community, died on 30 September at the age of 67 due to a heart attack. He was a violinist, violist, organist, and…

With the increasing global prevalence of chronic kidney disease (CKD), Maintenance Hemodialysis (MHD) has become one of the primary treatment modalities for patients with end-stage renal disease (ESRD).1 Although MHD plays a…
The contents of T3, C3, A3, G3, GC1, GC2, GC3, and GCall in cp coding gene codons of 49 Dendrobium species were analyzed in this study (Figure S1, Table S3). The results revealed an asymmetric expression of the four nucleotides in these codons. Specifically, the third position of codons showed a higher frequency of A/T usage compared to G/C.The nucleotide expression frequencies at the third codon position were as follows: T3 (45.72%−47.79%), A3 (40.92%−43.96%), G3 (16.68%−19.13%), and C3 (16.64%−19.51%). Analysis of GC content across the coding regions showed values of 42.14%−48.65% for GC1, 37.37%−41.08% for GC2, and 28.81%−32.14% for GC3, all of which were below 50%. This indicates an overall preference for codons rich in A/T nucleotides. Notably, the GC content exhibited the trend GC1 > GC2 > GC3, highlighting a stronger preference for codons ending with A/T in the cp coding genes of Dendrobium species.
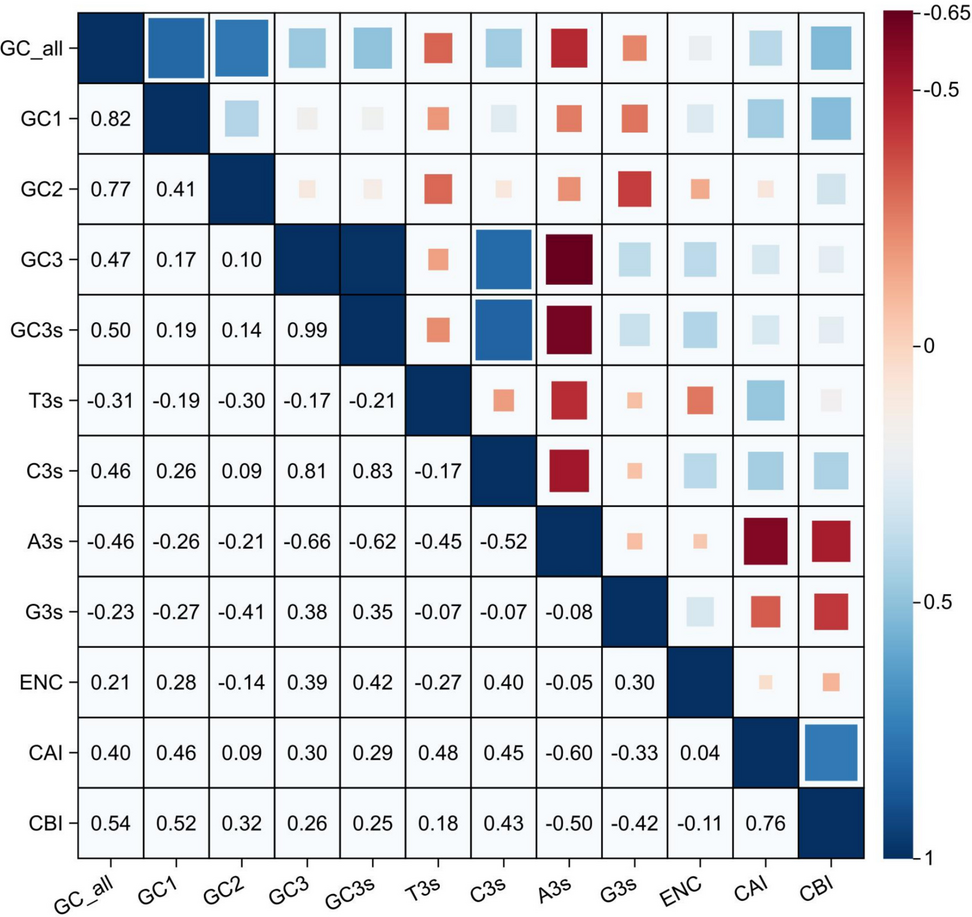
GC content is a critical indicator of genomic characteristics. we analyzed the correlation among GC1, GC2, GC3, and GCall (Fig. 1, Table S4). The results revealed that GCall was significantly correlated with GC1, GC2, and GC3 (P < 0.01). Furthermore, GC1 and GC2 were significantly correlated (P < 0.05), indicating that the GC composition at the first and second codon positions may influence each other. In contrast, GC3 showed no correlation with GC1 or GC2, suggesting that the GC composition at the third position is independent of the first and second positions.
Correlation analysis among the parameters of CUB (e.g., D. nobile). The numeric labels in the cells of the matrix represent correlation coefficients. These values typically range from -0.65 to 1. Dark blue indicates a positive correlation, while dark red indicates a negative correlation
RSCU is an important parameter for measuring codon preference. RSCU > 1 indicates a strong preference for that codon, categorizing as a high-frequency codon. Conversely, RSCU < 1 signifies a weak preference, classifying as a low-frequency codon. Excluding the initiation codon (AUG) and the termination codons (UGG, UAG, and UAA), there were 30 codons with RSCU > 1, representing 46.88% of the total number of codons (Fig. 2 and Table S5). Notably, 29 codons ended with A or U bases, accounting for 96.67% of the high-frequency codons. Among these, AGA, which encodes Arginine, exhibited the highest RSCU value, followed by GCU, which encodes Alanine. Additionally, 30 low-frequency codons with RSCU < 1 and 28 ended with G/C, accounting for 93.33%. These results indicated that the high-frequency codons tend to end with A/U, while the low-frequency codons are more likely to end with G/C.
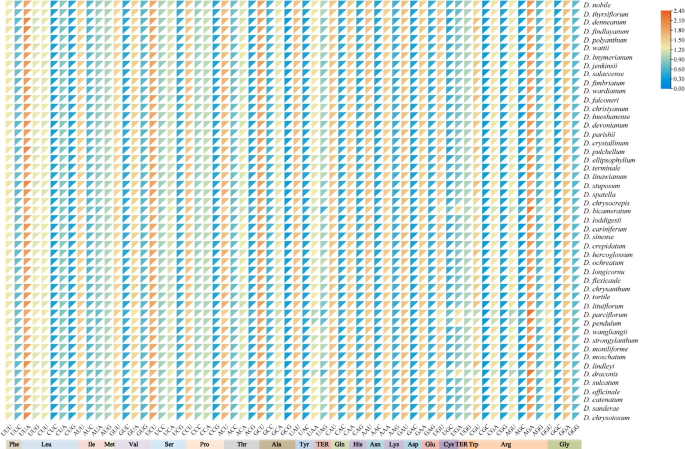
Heat Map for 49 Dendrobium species using RSCU values. The color and the degree of intensity indicate the RSCU value, and it varies from orange to blue with a low value to a high value of RSCU
A PR2-plot analysis was conducted to analyze the relationship between the third base of codons (A3/T3 and G3/C3) in CDS of cp genomes from 49 Dendrobium species. As shown in Fig. 3, cp coding genes in the four regions were unevenly distributed, with the majority located far from the center. This deviation suggests that CUB in these genes may be primarily governed by natural selection.
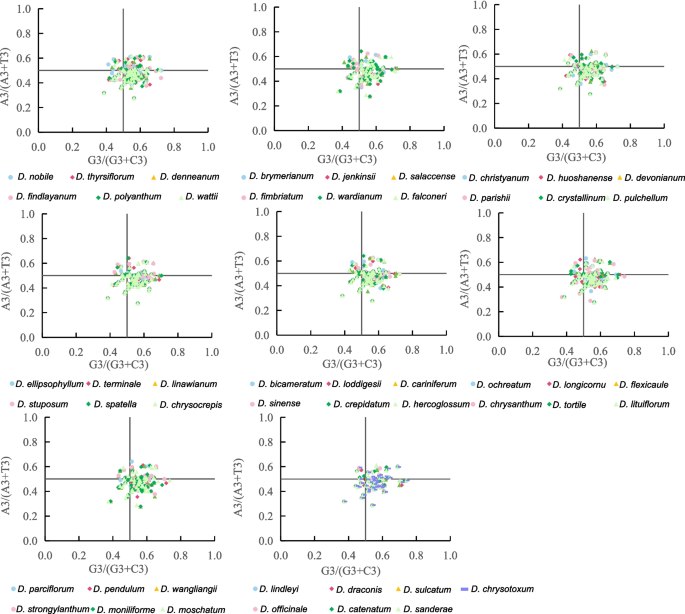
PR2-Plot of 49 Dendrobium species
To further explore the influence of natural selection and mutation pressure on CUB, neutrality plot analysis was conducted using GC12 and GC3 as lateral and vertical axes, respectively. The results showed that GC12 was distributed from 29.01% to 53.24%, while GC3 was distributed from 20.39% to 46.86% in the CDS of the cp genome (Fig. 4). Notably, few genes were distributed along the diagonal, indicating a weak correlation among the nucleotide compositions at GC12 and GC3. Furthermore, the regression coefficient (slope) ranged from −0.038 to 0.368.
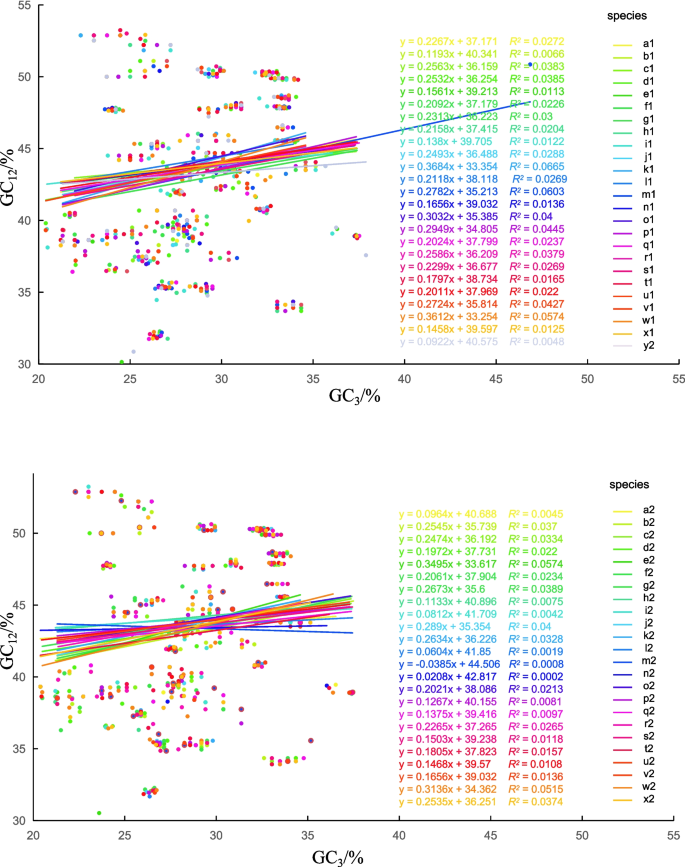
Neutrality plot analysis results of GC12 and GC3 content values. A: Lowercase letters (a1 to y1) indicate D. nobile (a1), D. thyrsiflorum (b1), D. denneanum (c1), D. findlayanum (d1), D. polyanthum (e1), D. wattii (f1), D. bnymerianum (g1), D. jenkinsii (h1), D. salaccense (i1), D. fimbriatum (j1), D. wardianum (k1), D. falconeri (l1), D. christyanum (m1), D. huoshanense (n1), D. devonianum (o1), D. parishii (p1), D. crystallinum (q1), D. pulchellum (r1), D. ellipsophyllum (s1), D. terminale (t1), D. linawianum (u1), D. stuposum (v1), D. spatella (w1), D. chrysocrepis (x1), D. bicameratum (y1) respectively.B: Lowercase letters (a2 to x2) indicate D. loddigesii (a2), D. cariniferum (b2), D. sinense (c2), D. crepidatum (d2), D. hercoglossum (e2), D. ochreatum (f2), D. longicornu (g2), D. flexicaule (h2), D. chrysanthum (i2), D. tortile (j2), D. lituiflorum (k2), D. parciflorum (l2), D. pendulum (m2), D. wangliangii (n2), D. strongylanthum (o2), D. moniliforme (p2), D. moschatum (q2), D. lindleyi (r2), D. draconis (s2), D. sulcatum (t2), D. officinale (u2), D. scabrilingue (v2), D. sanderae (w2), D. chrysotoxum (x2) respectively
An ENC-plot analysis for GC3 was performed to investigate the factors influencing CUB in CDS of Dendrobium cp genome. When the CUB is influenced only by mutation pressure, the cp genes will fall on the ENC expected curve. If the cp genes fall below the ENC expected curve, then the CUB is mainly driven by other factors such as natural selection, in addition to mutation pressure [38]. As shown in Fig. 5, the Majority of the cp genes in the 49 Dendrobium species were far from the expected ENC curve, indicating that natural selection may be the primary factor driving CUB in the cp genome of Dendrobium, while mutation pressure plays a secondary role.
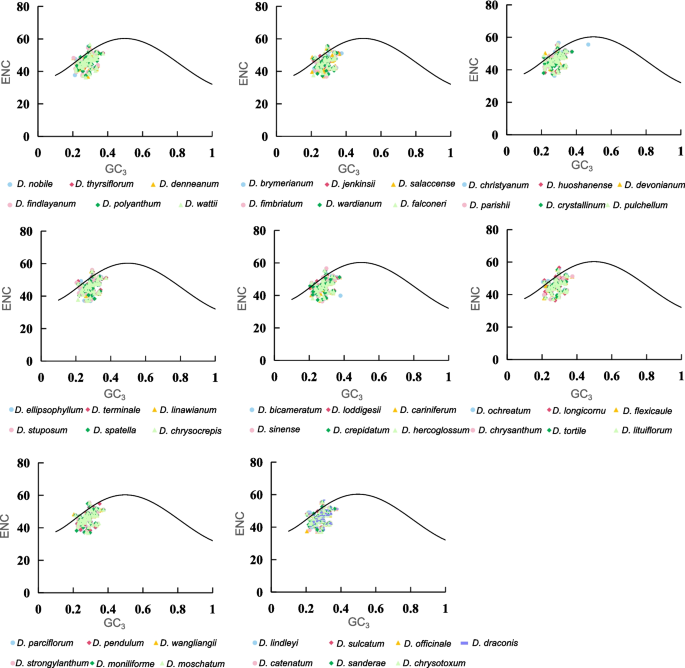
ENC-GC3 plots of 49 Dendrobium species
The calculated ENC ratios [(ENCexp-ENCobs)/ENCexp] range from −0.15 to 0.35 (Table 1), representing the difference between ENCobs values and ENCexp values. Approximately 73% of the genes have an ENC ratio greater than 0, which is consistent with the results of the ENC-plot analysis.
Eleven to nineteen optimal codons were identified across 49 Dendrobium species, with most optimal codons ending with A/U. Notably, D. polyanthum and D. devonianum shared the same optimal codons: GCA, GAA, UUU, GGA, CAU, UUA, AAU, CCU, AGA, CGA, AGU, GUA, and GUU. Similarly, D. nobile, D. linawianum, D. flexicaule, and D. officinale shared the same optimal codons: GCA, GAA, UUU, GGA, GGU, CAU, AUU, AAA, UUA, CCU, CAA, CGA, CGU, AGU, ACU, and UAU (Figure S2 and Table S6).
Phylogenetic trees of 49 Dendrobium cp genomes were constructed based on the cp genomics and CDS using the ML method, with C. hookerianum and C. lowianum as outgroups. The resulting phylogenetic topologies revealed that the Dendrobium species was divided into three distinct groups (Group A, Group B, and Group C), and some species showed overlap between the cp genomics and CDS phylogenetic trees (Fig. 6). Discrepancies were also observed between molecular biological evidence and traditional plant morphological classifications within individual clusters. Species in sct. Chrysotoxae is found across Groups A, B, and C, challenging traditional morphological classifications and suggesting that of sct. Chrysotoxae species may have maternal sources from sct. Dendrobium, sct. Holochrysa, and sct. Chrysotoxae. Similarly, D. hrysocrepis from sct. Holochrysa appeared in Group A, distinctly separated from Group B, indicating possible maternal lineage derived from sct. Dendrobium. D. hercoglossum as the sole representative from sct. Breviflores is more closely related to sct. Dendrobium. However, it is difficult to define the phylogenetic relationship between sct. Breviflores and sct. Dendrobium.
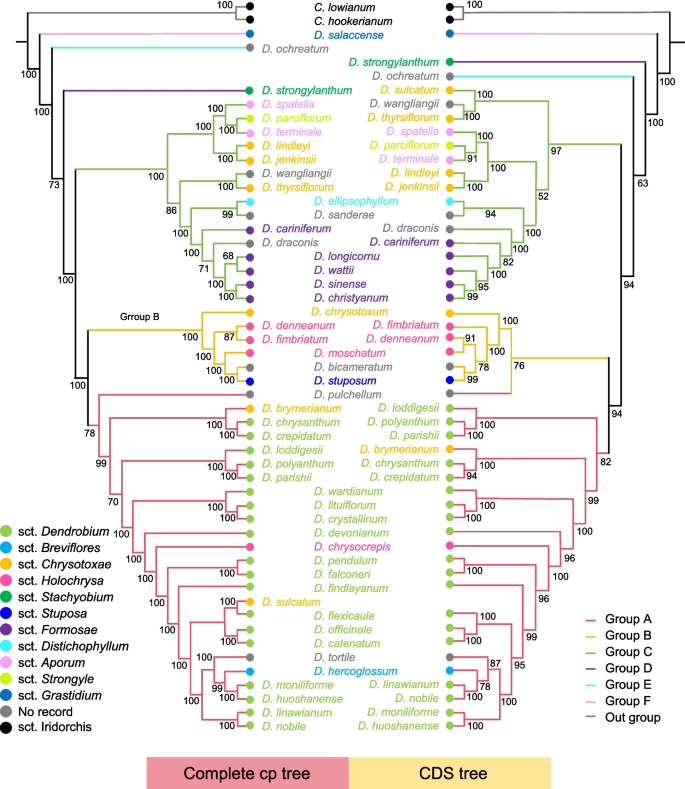
ML trees of 49 Dendrobium species using CDSs and cp genomes by the maximum likelihood method
Additionally, PLS-DA was conducted to evaluate the relationship between CUB and phylogeny based on RSCU values of Dendrobium species. The PLS-DA showed that Groups A and C were each divided into two subgroups, consistent with the phylogenetic trees derived from the cp genomics and CDS (Figure S3). In contrast, Group B could not form a distinct cluster. The species were dispersed into Groups A and C. These findings suggest that RSCU values are valuable tools for revealing the evolutionary relationships among Dendrobium species.

With immersive technology continuing to evolve, the intersection of neuroscience, AI, and XR is creating new possibilities for human well-being. In a forward-looking collaboration, Lenovo and Reulay are…
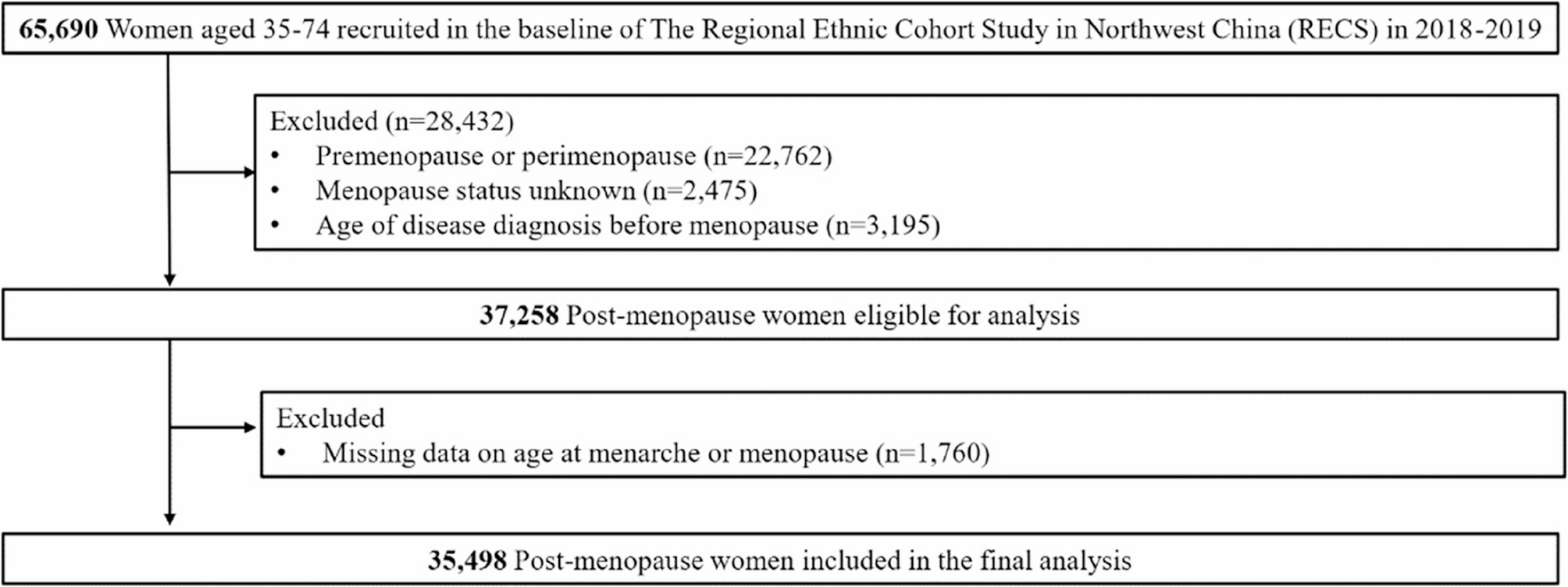
In this study, we observed a consistent inverse association of the duration of reproductive lifespan and lifetime endogenous estrogen exposure with the risks of diabetes, hypertension and CVD among postmenopausal women in Northwest China. Women…

Colton Herta will represent the Hitech squad when he switches from IndyCar to F2 in 2026, it has been announced, with the American attempting to pave the way for his “ultimate ambition” of one day racing in F1.
Herta, who was announced as a…