Global and regional disease burden of GCs
The global burden of GCs exhibits significant variation across cancer types, regions, and socio-economic contexts. Among these, BC remains the most prevalent and deadliest, with continued increases in…
The global burden of GCs exhibits significant variation across cancer types, regions, and socio-economic contexts. Among these, BC remains the most prevalent and deadliest, with continued increases in…
Starzl TE. History of clinical transplantation. World J Surg. 2000;24(7):759–82.
Google Scholar
Cooper JE. Evaluation and treatment of acute rejection in kidney allografts. Clin J Am Soc Nephrol CJASN. 2020;15(3):430–8.
Google Scholar
Lusco MA, Fogo AB, Najafian B, Alpers CE. AJKD atlas of renal pathology: acute T-cell–mediated rejection. Am J Kidney Dis. 2016;67(5):e29-30.
Google Scholar
Naesens M, Roufosse C, Haas M, Lefaucheur C, Mannon RB, Adam BA, et al. The Banff 2022 kidney meeting report: reappraisal of microvascular inflammation and the role of biopsy-based transcript diagnostics. Am J Transplant. 2024;24(3):338–49.
Google Scholar
Loupy A, Haas M, Roufosse C, Naesens M, Adam B, Afrouzian M, et al. The Banff 2019 kidney meeting report (I): updates on and clarification of criteria for T cell– and antibody-mediated rejection. Am J Transplant. 2020;20(9):2318–31.
Google Scholar
Nankivell BJ, Agrawal N, Sharma A, Taverniti A, P’Ng CH, Shingde M, et al. The clinical and pathological significance of borderline T cell–mediated rejection. Am J Transplant. 2019;19(5):1452–63.
Google Scholar
Loupy A, Mengel M, Haas M. Thirty years of the International banff classification for allograft pathology: the past, present, and future of kidney transplant diagnostics. Kidney Int. 2022;101(4):678–91.
Google Scholar
de Freitas DG, Sellarés J, Mengel M, Chang J, Hidalgo LG, Famulski KS, et al. The nature of biopsies with “borderline rejection” and prospects for eliminating this category. Am J Transplant. 2012;12(1):191–201.
Google Scholar
Mehta RB, Tandukar S, Jorgensen D, Randhawa P, Sood P, Puttarajappa C, et al. Early subclinical tubulitis and interstitial inflammation in kidney transplantation have adverse clinical implications. Kidney Int. 2020;98(2):436–47.
Google Scholar
Heilman RL, Nijim S, Chakkera HA, Devarapalli Y, Moss AA, Mulligan DC, et al. Impact of acute rejection on kidney allograft outcomes in recipients on rapid steroid withdrawal. J Transplant. 2011;2011(1):583981.
Google Scholar
Masin-Spasovska J, Spasovski G, Dzikova S, Petrusevska G, Dimova B, Lekovski L, et al. The evolution of untreated borderline and subclinical rejections at first month kidney allograft biopsy in comparison with histological changes at 6 months protocol biopsies. Prilozi. 2005;26(1):25–33.
Google Scholar
Thierry A, Thervet E, Vuiblet V, Goujon JM, Machet MC, Noel LH, et al. Long-term impact of subclinical inflammation diagnosed by protocol biopsy one year after renal transplantation. Am J Transplant. 2011;11(10):2153–61.
Google Scholar
Wang C, Feng G, Zhao J, Xu Y, Li Y, Wang L, et al. Screening of novel biomarkers for acute kidney transplant rejection using DIA-MS based proteomics. PROTEOMICS – Clinical Appl. 2024;18(3):2300047.
Heidari SS, Nafar M, Kalantari S, Tavilani H, Karimi J, Foster L, et al. Urinary epidermal growth factor is a novel biomarker for early diagnosis of antibody mediated kidney allograft rejection: A urinary proteomics analysis. J Proteomics. 2021;30(240):104208.
Gwinner W, Karch A, Braesen JH, Khalifa AA, Metzger J, Naesens M, et al. Noninvasive diagnosis of acute rejection in renal transplant patients using mass spectrometric analysis of urine samples: a multicenter diagnostic phase III trial. Transplant Direct. 2022;8(5):e1316.
Google Scholar
Geoui T, Urlaub H, Plessmann U, Porschewski P. Extraction of proteins from formalin-fixed, paraffin-embedded tissue using the qproteome extraction technique and preparation of tryptic peptides for liquid chromatography/mass spectrometry analysis. Current Protocls Mol Biol. 2010;90(1):10–27.
Wiśniewski JR, Gaugaz FZ. Fast and sensitive total protein and Peptide assays for proteomic analysis. Anal Chem. 2015;87(8):4110–6.
Google Scholar
Conover WJ. Practical nonparametric statistics. 3rd ed. New York: Wiley; 1998.
Jonckheere AR. A distribution-free k-sample test against ordered alternatives. Biometrika. 1954;41(1–2):133–45.
Pallant J. SPSS Survival Manual: A Step by Step Guide to Data Analysis Using SPSS for Windows. 4th ed. McGraw Hill Open University Press; 2010.
Cohen J. Statistical Power Analysis for the Behavioral Sciences. 2nd ed. Hillsdale; 1988.
Armitage P. Tests for Linear Trends in Proportions and Frequencies. Biometrics. 1955;11(3):375.
Fang F, Liu P, Song L, Wagner P, Bartlett D, Ma L, et al. Diagnosis of T-cell-mediated kidney rejection by biopsy-based proteomic biomarkers and machine learning. Front Immunol. 2023;6(14):1090373.
Hofstraat R, Marx K, Blatnik R, Claessen N, Chojnacka A, Peters-Sengers H, et al. Highly Repeatable Tissue Proteomics for Kidney Transplant Pathology: Technical and Biological Validation of Protein Analysis using LC-MS/MS. bioRxiv; 2024. p 2024.06.14.599091.
Schmouder RL, Kunkel SL. The cytokine response in renal allograft rejection. Nephrol Dial Transplant. 1995;10(supp1):36–43.
Google Scholar
De Serres SA, Mfarrej BG, Grafals M, Riella LV, Magee CN, Yeung MY, et al. Derivation and validation of a cytokine-based assay to screen for acute rejection in renal transplant recipients. Clin J Am Soc Nephrol. 2012;7(6):1018.
Google Scholar
Karczewski J, Karczewski M, Glyda M, Wiktorowicz K. Role of TH1/TH2 cytokines in kidney allograft rejection. Transplant Proc. 2008;40(10):3390–2.
Google Scholar
Biro M, Munoz MA, Weninger W. Targeting Rho-GTPases in immune cell migration and inflammation. Br J Pharmacol. 2014;171(24):5491–506.
Google Scholar
Matsuda J, Asano-Matsuda K, Kitzler TM, Takano T. Rho GTPase regulatory proteins in podocytes. Kidney Int. 2021;99(2):336–45.
Google Scholar
Kurian SM, Velazquez E, Thompson R, Whisenant T, Rose S, Riley N, et al. Orthogonal comparison of molecular signatures of kidney transplants with subclinical and clinical acute rejection: equivalent performance is agnostic to both technology and platform. Am J Transplant. 2017;17(8):2103–16.
Google Scholar
Kurian SM, Williams AN, Gelbart T, Campbell D, Mondala TS, Head SR, et al. Molecular classifiers for acute kidney transplant rejection in peripheral blood by whole genome gene expression profiling. Am J Transplant. 2014;14(5):1164–72.
Google Scholar
Zhang M, Chen F, Feng S, Liu X, Wang Z, Shen N, et al. FBLN5 as one presumably prognostic gene potentially modulating tumor immune microenvironment for renal clear cell carcinoma in children and young adults. Pharmacogenomics Pers Med. 2024;19(17):27–40.
Huang W, Shi H, Hou Q, Mo Z, Xie X. GSTM1 and GSTT1 polymorphisms contribute to renal cell carcinoma risk: evidence from an updated meta-analysis. Sci Rep. 2015;5(1):17971.
Google Scholar
Li P, Li D, Lu Y, Pan S, Cheng F, Li S, et al. GSTT1/GSTM1 deficiency aggravated cisplatin-induced acute kidney injury via ROS-triggered ferroptosis. Front Immunol. 2024;25(15):1457230.
Beothe T, Docs J, Kovacs G, Peterfi L. Increased level of TXNIP and nuclear translocation of TXN is associated with end stage renal disease and development of multiplex renal tumours. BMC Nephrol. 2024;17(25):227.
Luczak M, Formanowicz D, Marczak Ł, Pawliczak E, Wanic-Kossowska M, Figlerowicz M, et al. Deeper insight into chronic kidney disease-related atherosclerosis: comparative proteomic studies of blood plasma using 2DE and mass spectrometry. J Transl Med. 2015;13(1):20.
Google Scholar
Abdullah EL, Jalalonmuhali M, Ng KP, Jamaluddin FA, Lim SK. The role of lymphocyte subset in predicting allograft rejections in kidney transplant recipients. Transplant Proceed. 2022;54(2):312–9.
Halloran PF. T cell-mediated rejection of kidney transplants: a personal viewpoint. Am J Transplant. 2010;10(5):1126–34.
Google Scholar
Liu B, Chen L, Huang H, Huang H, Jin H, Fu C. Prognostic and immunological value of GNB4 in gastric cancer by analyzing TCGA database. Dis Markers. 2022;16(2022):1–16.
Zhan J, Huang L, Niu L, Lu W, Sun C, Liu S, et al. Regulation of CD73 on NAD metabolism: unravelling the interplay between tumour immunity and tumour metabolism. Cell Commun Signal. 2024;22(1):387.
Google Scholar
Chen S, Wainwright DA, Wu JD, Wan Y, Matei DE, Zhang Y, et al. Cd73: an emerging checkpoint for cancer immunotherapy. Immunotherapy. 2019;11(11):983–97.
Google Scholar
Antonioli L, Pacher P, Vizi ES, Haskó G. CD39 and CD73 in immunity and inflammation. Trends Mol Med. 2013;19(6):355–67.
Google Scholar
Schneider E, Winzer R, Rissiek A, Ricklefs I, Meyer-Schwesinger C, Ricklefs FL, et al. CD73-mediated adenosine production by CD8 T cell-derived extracellular vesicles constitutes an intrinsic mechanism of immune suppression. Nat Commun. 2021;12(1):5911.
Google Scholar
Deutsch EW, Bandeira N, Perez-Riverol Y, Sharma V, Carver JJ, Mendoza L, et al. The proteomexchange consortium at 10 years: 2023 update. Nucleic Acids Res. 2023;51(D1):D1539–48.
Google Scholar
Perez-Riverol Y, Bandla C, Kundu DJ, Kamatchinathan S, Bai J, Hewapathirana S, et al. The PRIDE database at 20 years: 2025 update. Nucleic Acids Res. 2025;53(D1):D543–53.
Google Scholar

Mosquitoes have been found in Iceland for the first time as global heating makes the country more hospitable for insects.
The country was until this month one of the only places in the world that did not have a mosquito population. The other is…

Read more news stories here
Dr Louis van der Watt, a well-respected string teacher in the South African classical music community, died on 30 September at the age of 67 due to a heart attack. He was a violinist, violist, organist, and…

With the increasing global prevalence of chronic kidney disease (CKD), Maintenance Hemodialysis (MHD) has become one of the primary treatment modalities for patients with end-stage renal disease (ESRD).1 Although MHD plays a…
The contents of T3, C3, A3, G3, GC1, GC2, GC3, and GCall in cp coding gene codons of 49 Dendrobium species were analyzed in this study (Figure S1, Table S3). The results revealed an asymmetric expression of the four nucleotides in these codons. Specifically, the third position of codons showed a higher frequency of A/T usage compared to G/C.The nucleotide expression frequencies at the third codon position were as follows: T3 (45.72%−47.79%), A3 (40.92%−43.96%), G3 (16.68%−19.13%), and C3 (16.64%−19.51%). Analysis of GC content across the coding regions showed values of 42.14%−48.65% for GC1, 37.37%−41.08% for GC2, and 28.81%−32.14% for GC3, all of which were below 50%. This indicates an overall preference for codons rich in A/T nucleotides. Notably, the GC content exhibited the trend GC1 > GC2 > GC3, highlighting a stronger preference for codons ending with A/T in the cp coding genes of Dendrobium species.
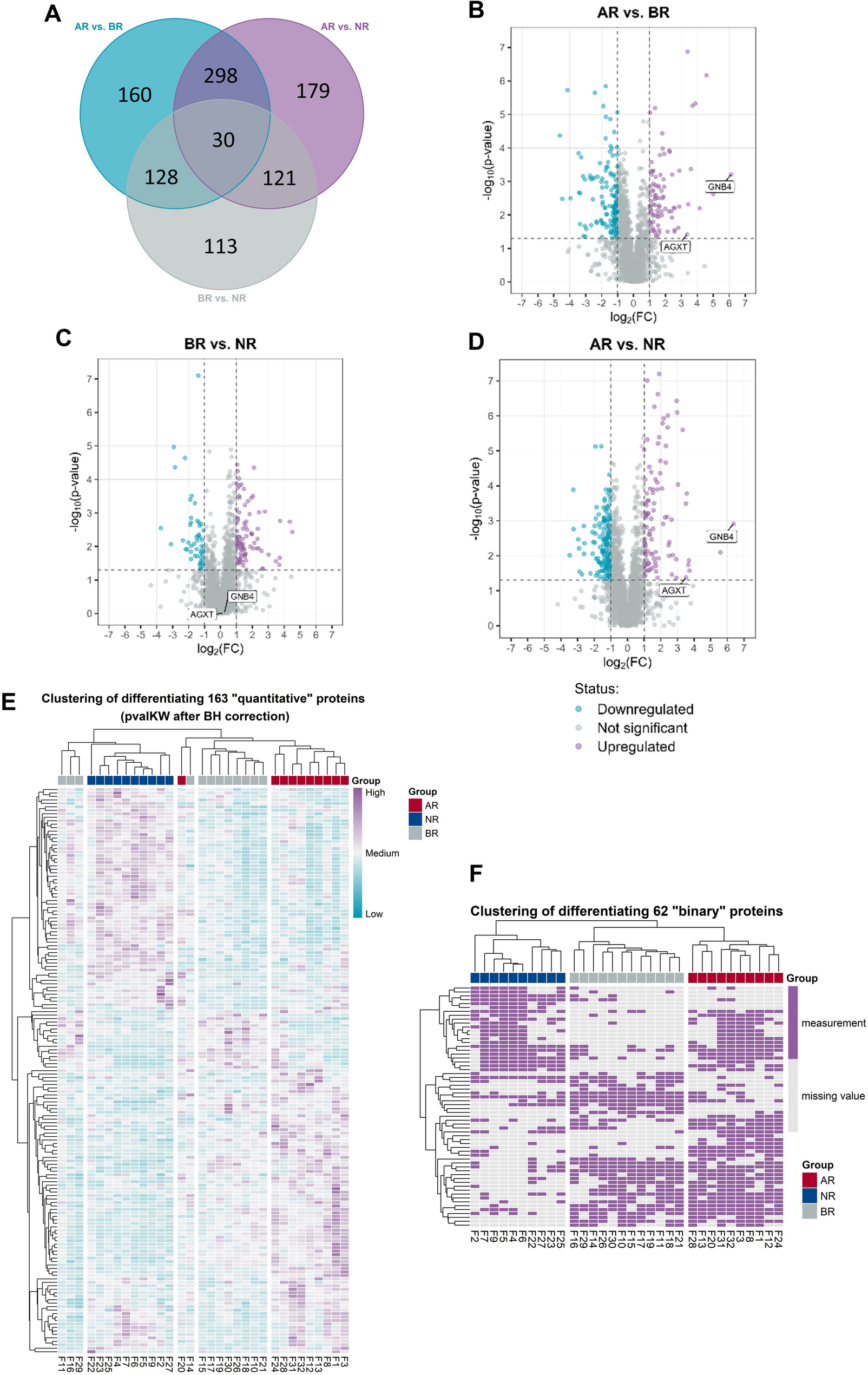
GC content is a critical indicator of genomic characteristics. we analyzed the correlation among GC1, GC2, GC3, and GCall (Fig. 1, Table S4). The results revealed that GCall was significantly correlated with GC1, GC2, and GC3 (P < 0.01). Furthermore, GC1 and GC2 were significantly correlated (P < 0.05), indicating that the GC composition at the first and second codon positions may influence each other. In contrast, GC3 showed no correlation with GC1 or GC2, suggesting that the GC composition at the third position is independent of the first and second positions.
Correlation analysis among the parameters of CUB (e.g., D. nobile). The numeric labels in the cells of the matrix represent correlation coefficients. These values typically range from -0.65 to 1. Dark blue indicates a positive correlation, while dark red indicates a negative correlation
RSCU is an important parameter for measuring codon preference. RSCU > 1 indicates a strong preference for that codon, categorizing as a high-frequency codon. Conversely, RSCU < 1 signifies a weak preference, classifying as a low-frequency codon. Excluding the initiation codon (AUG) and the termination codons (UGG, UAG, and UAA), there were 30 codons with RSCU > 1, representing 46.88% of the total number of codons (Fig. 2 and Table S5). Notably, 29 codons ended with A or U bases, accounting for 96.67% of the high-frequency codons. Among these, AGA, which encodes Arginine, exhibited the highest RSCU value, followed by GCU, which encodes Alanine. Additionally, 30 low-frequency codons with RSCU < 1 and 28 ended with G/C, accounting for 93.33%. These results indicated that the high-frequency codons tend to end with A/U, while the low-frequency codons are more likely to end with G/C.
Heat Map for 49 Dendrobium species using RSCU values. The color and the degree of intensity indicate the RSCU value, and it varies from orange to blue with a low value to a high value of RSCU
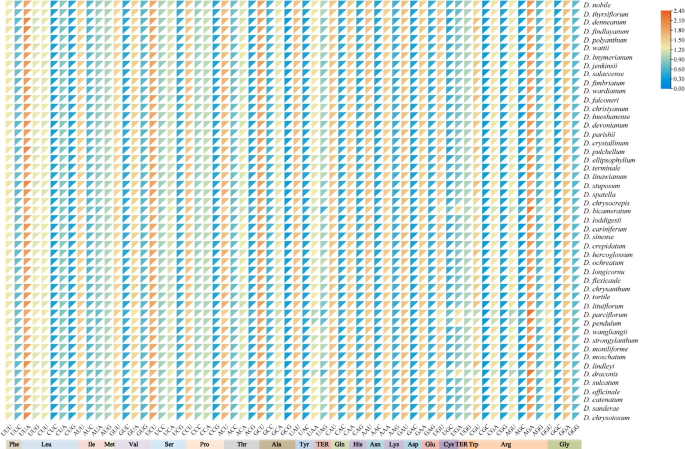
A PR2-plot analysis was conducted to analyze the relationship between the third base of codons (A3/T3 and G3/C3) in CDS of cp genomes from 49 Dendrobium species. As shown in Fig. 3, cp coding genes in the four regions were unevenly distributed, with the majority located far from the center. This deviation suggests that CUB in these genes may be primarily governed by natural selection.
PR2-Plot of 49 Dendrobium species
To further explore the influence of natural selection and mutation pressure on CUB, neutrality plot analysis was conducted using GC12 and GC3 as lateral and vertical axes, respectively. The results showed that GC12 was distributed from 29.01% to 53.24%, while GC3 was distributed from 20.39% to 46.86% in the CDS of the cp genome (Fig. 4). Notably, few genes were distributed along the diagonal, indicating a weak correlation among the nucleotide compositions at GC12 and GC3. Furthermore, the regression coefficient (slope) ranged from −0.038 to 0.368.
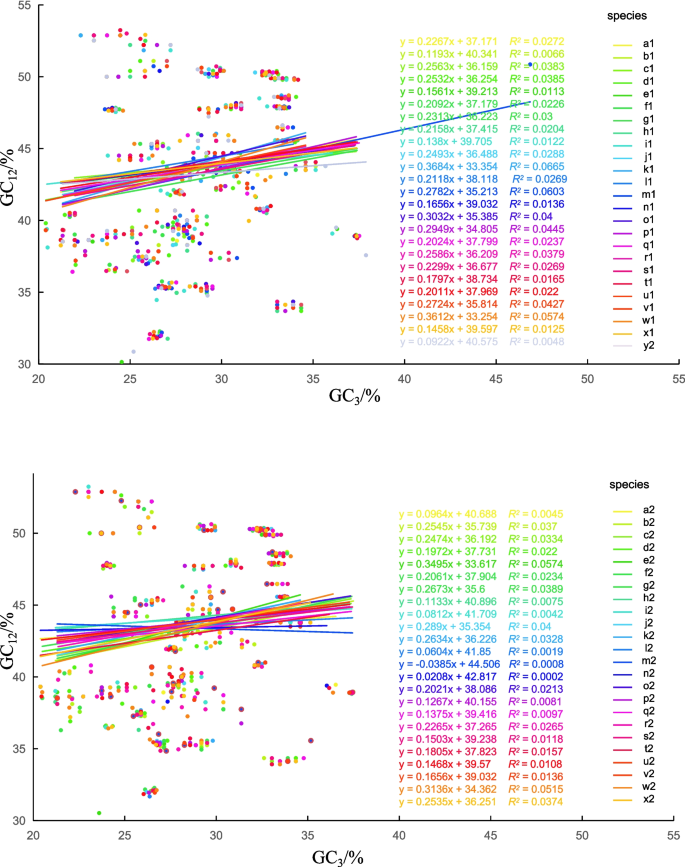
Neutrality plot analysis results of GC12 and GC3 content values. A: Lowercase letters (a1 to y1) indicate D. nobile (a1), D. thyrsiflorum (b1), D. denneanum (c1), D. findlayanum (d1), D. polyanthum (e1), D. wattii (f1), D. bnymerianum (g1), D. jenkinsii (h1), D. salaccense (i1), D. fimbriatum (j1), D. wardianum (k1), D. falconeri (l1), D. christyanum (m1), D. huoshanense (n1), D. devonianum (o1), D. parishii (p1), D. crystallinum (q1), D. pulchellum (r1), D. ellipsophyllum (s1), D. terminale (t1), D. linawianum (u1), D. stuposum (v1), D. spatella (w1), D. chrysocrepis (x1), D. bicameratum (y1) respectively.B: Lowercase letters (a2 to x2) indicate D. loddigesii (a2), D. cariniferum (b2), D. sinense (c2), D. crepidatum (d2), D. hercoglossum (e2), D. ochreatum (f2), D. longicornu (g2), D. flexicaule (h2), D. chrysanthum (i2), D. tortile (j2), D. lituiflorum (k2), D. parciflorum (l2), D. pendulum (m2), D. wangliangii (n2), D. strongylanthum (o2), D. moniliforme (p2), D. moschatum (q2), D. lindleyi (r2), D. draconis (s2), D. sulcatum (t2), D. officinale (u2), D. scabrilingue (v2), D. sanderae (w2), D. chrysotoxum (x2) respectively
An ENC-plot analysis for GC3 was performed to investigate the factors influencing CUB in CDS of Dendrobium cp genome. When the CUB is influenced only by mutation pressure, the cp genes will fall on the ENC expected curve. If the cp genes fall below the ENC expected curve, then the CUB is mainly driven by other factors such as natural selection, in addition to mutation pressure [38]. As shown in Fig. 5, the Majority of the cp genes in the 49 Dendrobium species were far from the expected ENC curve, indicating that natural selection may be the primary factor driving CUB in the cp genome of Dendrobium, while mutation pressure plays a secondary role.
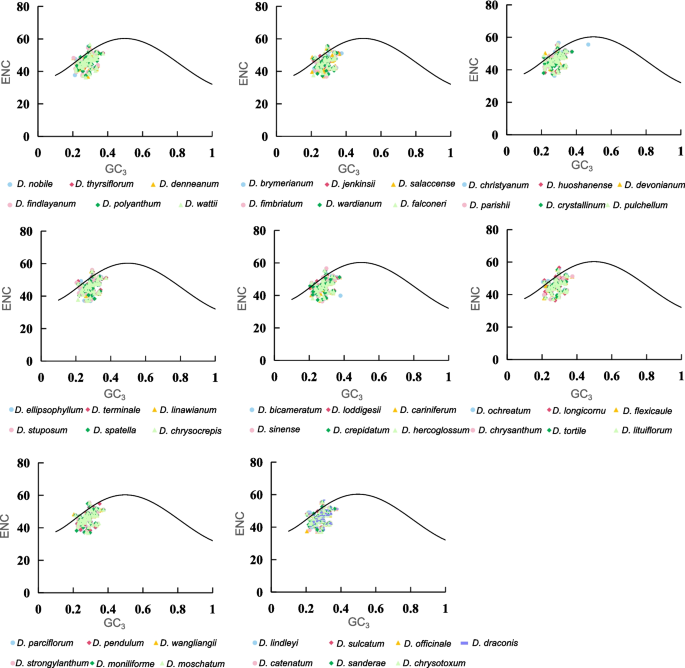
ENC-GC3 plots of 49 Dendrobium species
The calculated ENC ratios [(ENCexp-ENCobs)/ENCexp] range from −0.15 to 0.35 (Table 1), representing the difference between ENCobs values and ENCexp values. Approximately 73% of the genes have an ENC ratio greater than 0, which is consistent with the results of the ENC-plot analysis.
Eleven to nineteen optimal codons were identified across 49 Dendrobium species, with most optimal codons ending with A/U. Notably, D. polyanthum and D. devonianum shared the same optimal codons: GCA, GAA, UUU, GGA, CAU, UUA, AAU, CCU, AGA, CGA, AGU, GUA, and GUU. Similarly, D. nobile, D. linawianum, D. flexicaule, and D. officinale shared the same optimal codons: GCA, GAA, UUU, GGA, GGU, CAU, AUU, AAA, UUA, CCU, CAA, CGA, CGU, AGU, ACU, and UAU (Figure S2 and Table S6).
Phylogenetic trees of 49 Dendrobium cp genomes were constructed based on the cp genomics and CDS using the ML method, with C. hookerianum and C. lowianum as outgroups. The resulting phylogenetic topologies revealed that the Dendrobium species was divided into three distinct groups (Group A, Group B, and Group C), and some species showed overlap between the cp genomics and CDS phylogenetic trees (Fig. 6). Discrepancies were also observed between molecular biological evidence and traditional plant morphological classifications within individual clusters. Species in sct. Chrysotoxae is found across Groups A, B, and C, challenging traditional morphological classifications and suggesting that of sct. Chrysotoxae species may have maternal sources from sct. Dendrobium, sct. Holochrysa, and sct. Chrysotoxae. Similarly, D. hrysocrepis from sct. Holochrysa appeared in Group A, distinctly separated from Group B, indicating possible maternal lineage derived from sct. Dendrobium. D. hercoglossum as the sole representative from sct. Breviflores is more closely related to sct. Dendrobium. However, it is difficult to define the phylogenetic relationship between sct. Breviflores and sct. Dendrobium.
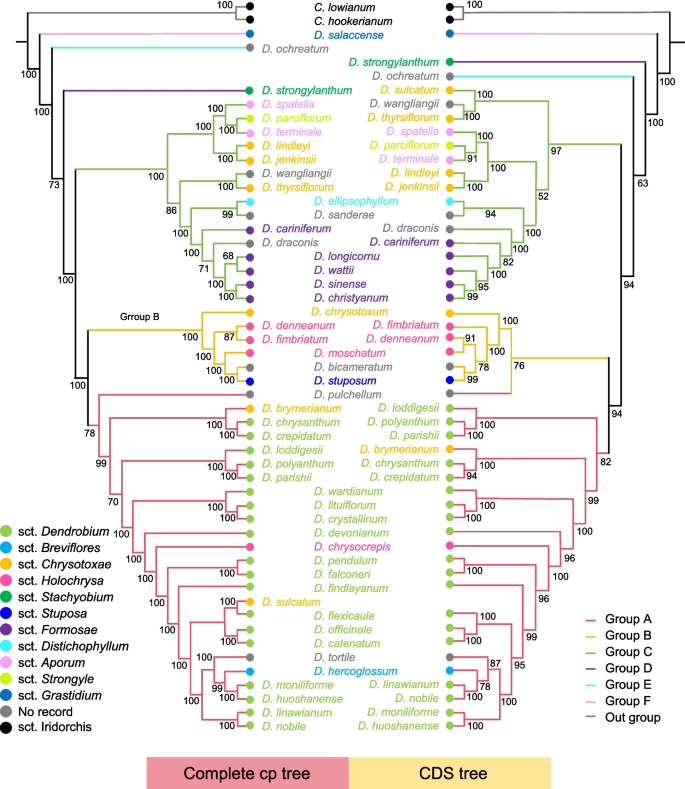
ML trees of 49 Dendrobium species using CDSs and cp genomes by the maximum likelihood method
Additionally, PLS-DA was conducted to evaluate the relationship between CUB and phylogeny based on RSCU values of Dendrobium species. The PLS-DA showed that Groups A and C were each divided into two subgroups, consistent with the phylogenetic trees derived from the cp genomics and CDS (Figure S3). In contrast, Group B could not form a distinct cluster. The species were dispersed into Groups A and C. These findings suggest that RSCU values are valuable tools for revealing the evolutionary relationships among Dendrobium species.

With immersive technology continuing to evolve, the intersection of neuroscience, AI, and XR is creating new possibilities for human well-being. In a forward-looking collaboration, Lenovo and Reulay are…
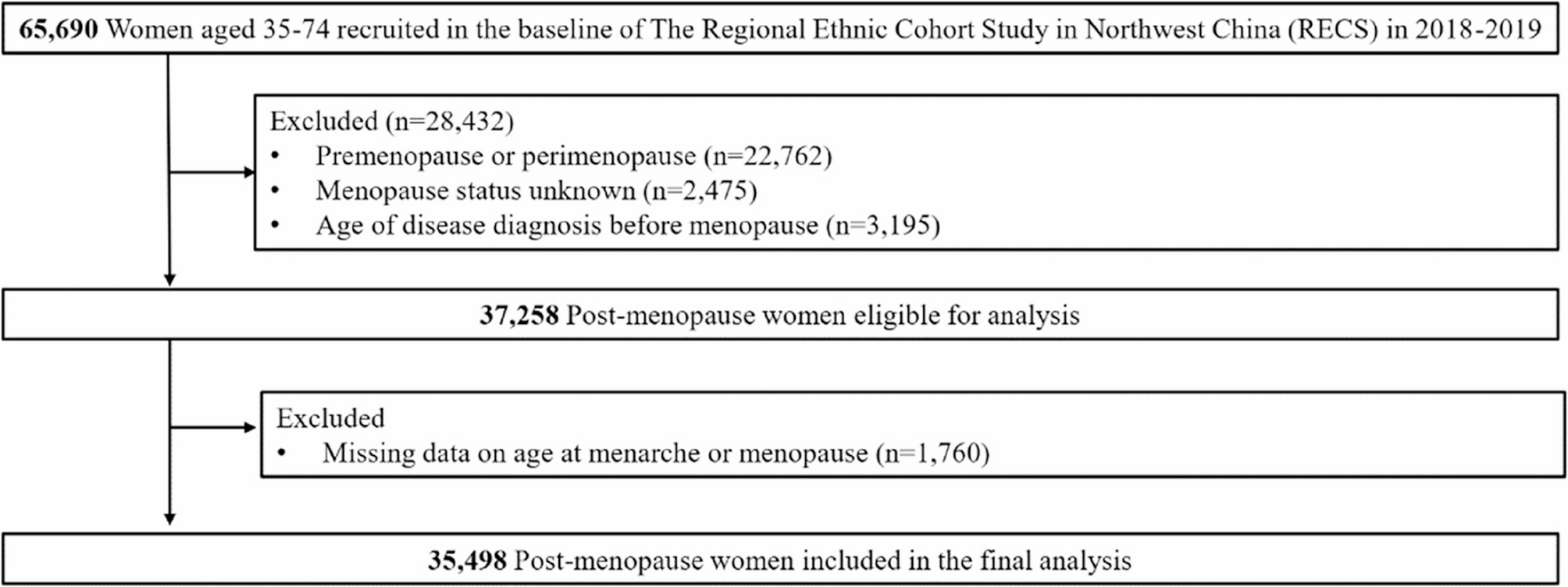
In this study, we observed a consistent inverse association of the duration of reproductive lifespan and lifetime endogenous estrogen exposure with the risks of diabetes, hypertension and CVD among postmenopausal women in Northwest China. Women…