UBC Okanagan doctoral student Tuan-Anh Nguyen, left, and Dr. Thu-Thuy Dang examine plant samples in their lab. Their research has uncovered how tropical trees produce mitraphylline, a rare compound with potential anti-tumour…
Blog
-

Instagram to bring in version of PG-13 system to protect children, says Meta | Instagram
Instagram is to adopt a version of the PG-13 cinema rating system to give parents stronger controls over their teenagers’ use of the social media platform.
Instagram, which is run by Meta, will start applying rules similar to the US “parental…
Continue Reading
-

Nicholas Sparks on Collaborating With M. Night Shyamalan for ‘Remain’
Nicholas Sparks and M. Night Shyamalan are more similar than you think. And though joining forces for a novel may seem like a unique choice, the pair’s paths have crossed before.
“Long time ago, when we had the original script of The…
Continue Reading
-

Fibocom Leads the FWA Evolution at NetworkX 2025 with AI-Powered Connectivity Innovations
PARIS, Oct. 14, 2025 /PRNewswire/ — Fibocom, a leading global provider of wireless communication modules and AI solutions, is showcasing its leadership in 5G Fixed Wireless…
Continue Reading
-
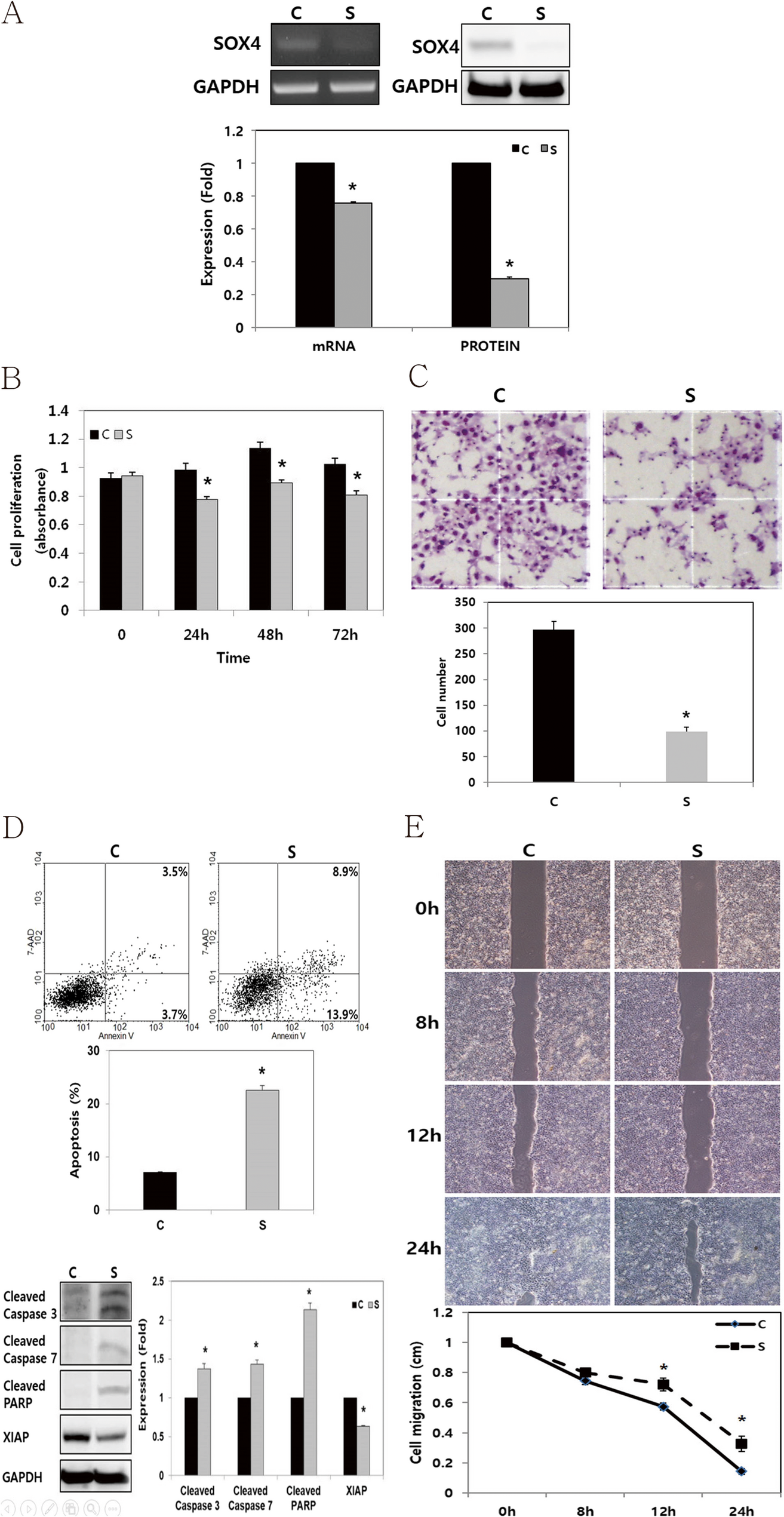
SOX4 enhances tumor progression and cisplatin resistance in orthotopic mouse xenograft model of head and neck squamous cell carcinoma | BMC Cancer
Cell culture and transfection
The HNSCC, FADU cell lines were purchased from the American Type Culture Collection (Manassas, VA, USA). FADU cell line were cultured in DMEM/F12 medium (GIBCO®, Invitrogen, Carlsbad, CA, USA) with 10% fetal bovine serum (FBS, GIBCO®, Invitrogen) and 1% penicillin/streptomycin. Cells were cultured in a Humidified incubator at 37°C with 5% CO2. To knock down endogenous SOX4 gene expression in FADU cells, small interfering RNAs (siRNAs) were utilized. FADU cells were seeded into 6-well plates at a density of 2.0 × 105 cells/well and transfected with a SOX4-specific (Bioneer, Daejeon, Korea) or a negative control siRNA (cat. no. 1027281, Qiagen, Germantown, MD, USA) using (Invitrogen) for 48 h at 37°C. The SOX4-specific si-RNA sequences were as follows: Sense, 5’- GAU AGA UGG CGC UAU CUU U-3’ and Antisense, 5’-AAA CAU AGC GCC AUC UAU C −3’. Subsequent analyses were conducted 48 h post-transfection.
RNA isolation and reverse-transcription polymerase chain reaction
Total RNA was extracted from FADU using TRIzol reagent (Invitrogen) following the manufacturer’s protocol. For reverse transcription (RT), total RNA (1 µg), M-MLV reverse transcriptase (Invitrogen), 1 µL of 2 mM dNTP mix (Enzynomics Co., Ltd., Daejeon, Korea), 2 µL of 0.1 M dithiothreitol (Invitrogen), 4 µL of 5× first strand buffer (Invitrogen), 1 µL of RNase inhibitor (Promega Corporation), and 1 µL of oligo dT (Bioneer Corporation, Daejeon, Korea) were used. Primers specific for SOX4 and Glyceraldehyde-3-Phosphate Dehydrogenase (GAPDH, Bioneer Corporation) were utilized to amplify the cDNA. Polymerase chain reaction (PCR) was performed using GoTaq DNA Polymerase and 5× Green GoTaq reaction buffer (Promega Corporation). The primer sequences employed were as follows: SOX4 forward, 5ʹ- GCA CAT GGC TGA CTA CCC C-3ʹ; SOX4 reverse, 5ʹ- GCC TTG TAC AGC GAG TGG TG-3ʹ; GAPDH forward, 5ʹ-ACC ACA GTC CAT GCC ATC AC-3ʹ; and GAPDH reverse, 5ʹ-TCC ACC CTG TTG CTG TA-3ʹ. PCR products were separated via electrophoresis on a 1% agarose gel containing ethidium bromide.
Protein isolation and western blot analysis
Cells were lysed using a radioimmunoprecipitation assay buffer (Biosesang Inc.). Protein concentrations were subsequently measured using a bicinchoninic acid assay. Protein lysates (20–30 µg per lane) were separated via sodium dodecyl sulfate-polyacrylamide gel electrophoresis on 10%–12% gels and electrophoretically transferred onto polyvinylidene fluoride membranes. The membranes were then incubated at room temperature for 1 h with 5% bovine serum albumin (BSA; Bioshop Canada Inc.) in Tris-buffered saline (TBS) containing 0.1% Tween-20. The membranes were then washed four times for 15 min each with TBS–0.1% Tween-20. Specific proteins were identified using primary antibodies targeting GAPDH (cat. no. sc-25778; Santa Cruz Biotechnology, Inc., Dallas, TX, USA), β-actin (cat. no. 3700 S; Cell signaling Technology, Inc.), SOX4 (cat. no. ab80261; Abcam, UK), Cleaved caspase-3 (cat. no. 9664; Cell Signaling Technology, Inc.), Cleaved caspase-7 (cat. no. 9491; Cell Signaling Technology, Inc.), X-linked inhibitor of apoptosis protein (XIAP, cat.no. sc-11426; Santa Cruze Biotechnology, Texas, USA), and Cleaved poly (ADP-ribose) polymerase (PARP; cat. no. 5625; Cell Signaling Technology, Inc.; cat. no. ab32064; Abcam). The primary antibodies were diluted at a 1:1,000 ratios in TBS–0.1% Tween-20 and incubated with the membranes for 24 h at 4 °C. Secondary antibodies, anti-rabbit (cat. no. 7074; Cell Signaling Technology, Inc.) or anti-mouse (cat. no. 7076; Cell Signaling Technology, Inc.), were diluted at 1:2,000 ratio, and incubated at room temperature for 1 h. Immunoreactive proteins were visualized using a LAS 4000 luminescence image analyzer (FUJIFILM Wako Pure Chemical Corporation) and an enhanced chemiluminescence detection system for HRP (EMD Millipore). Western blot analysis was independently conducted in triplicate.
Cell proliferation assay
Following of transfection, cells plated in 24-well plates (1 × 104 cells per well). After an additional 48 h incubation, cell viability was measured using an EZ-CyTox (WST-1) enhanced cell viability assay kit (cat. no. EZ-3000; Daeil Lab Inc.) at 37 °C for 1–2 h. Absorbance was measured at 460 nm using a microplate reader. Cell viability assays were performed in triplicate and repeated independently.
Cell invasion assay
The extent of cell invasion was evaluated by counting the number of cells that migrated through an 8.0 μm pore Transwell invasion chamber (cat. no. 3422; Costar, Inc.). The upper chamber was coated with a 1% gelatin solution and incubated for 12 h at 37 °C, followed by 12 h of drying at room temperature before the experiment. After 48 h of transfection, cells in the upper chamber were seeded at a density of 2 × 105 cells in 120 µl of 0.2% BSA (BioShop Canada, Inc.) in FBS-free DMEM. As a chemoattractant, 400 µl of 0.2% BSA in FBS-free DMEM containing fibronectin (cat. no.361635; Calbiochem, San Diego, CA, USA) was loaded into the lower chamber. After 24 h of incubation, cells that had migrated to the bottom of the Transwell membranes were stained with Diff-Quik solution (Sysmex Corporation). Using a Light microscope, the cells were then counted in five random fields at 100× magnification. Results were expressed as mean ± standard error of the number of cells per field from three independent experiments.
Cell migration assay (wound healing assay)
The cells were seeded into each well of Culture-Inserts (Ibidi GmbH) at 1.5 × 105 cells/well, following transfection. They were incubated for 24 h. Following that, each insert was detached, and the progression of cell migration was assessed by imaging at 0, 8, 12, and 24 h using an inverted microscope. Distances between gaps were normalized to 1 cm after three random sites were captured.
Apoptosis assay
Apoptosis was evaluated using an APC Annexin V assay. Following transfection, the cells were harvested via trypsinization, washed twice with phosphate-buffered saline, and resuspended in a binding buffer (cat. no. 556454; BD Biosciences, San Jose, CA, USA). After adding APC Annexin V (cat. no. 550474) and 7-amino-actinomycin D (cat. no. 559925; BD Biosciences, San Jose, CA, USA), the cells were incubated in the dark for 15 min. The samples were subsequently resuspended in 400 µl of binding buffer. They were analyzed using a FACSCalibur flow cytometer (BD Biosciences) and BD Cell Quest version 3.3 software (Becton Dickinson). Data analysis was performed using WinMDI version 2.9 (The Scripps Research Institute). Apoptosis assays were conducted independently in triplicate.
Cell irradiation or cisplatin treatment
After 48 h of transfection, the cells were maintained at 37 °C and exposed to γ-irradiation at varying doses (8–10 Gy, 137Cs, and 2.875 Gy/min) using a Gammacell 3000 Elan (Therathronics) at room temperature. A stock solution of cisplatin (10 mg/20 ml; Dong-A, Co., Ltd., Seoul, Korea) was prepared and diluted to 2.5 ~ 5 µg/ml concentrations. The cisplatin solution was incubated at 37 °C for 24 h before being used in experiments.
Establishment of SOX4 overexpressing stable cell line and an orthotopic mouse xenograft model of head and neck squamous cell carcinoma
A stable SOX4-overexpressing SCC VII mouse squamous cell carcinoma cell line was established. After transfection of pcDNA6/myc-SOX4 into SCC VII cells using Lipofectamine 2000 (Thermo Fisher Scientific, USA), transfected cells were selected by culturing in media containing blasticidin (cat. no. A11139-03; ThermoFisher, Massachusetts, USA) at 5 µg/ml concentration. The stable overexpression of SOX4 in selected clones was confirmed via Western blot analysis. Female C3H/HeJ syngeneic mice (6–8 weeks old) were purchased from OrientBio (Seongnam, South Korea), and randomly assigned to a control or SOX4 overexpression group. Furthermore, 1 × 106 of SOX4 overexpressed or controlled SCC VII cells were suspended in 70 µl of serum-free media and slowly injected into the floor of the mouth (FOM) of mice via an intraoral approach. These animal experimental procedures were approved by the Chonnam National University Animal Care and Use Committee (CNU IACUC-H-201624). All animal care, experiments and euthanasia were performed per protocols approved by the Chonnam National University Animal Research Committee. For the euthanasia of mice, mice were first rendered unconscious with isoflurane, and then death was confirmed after 5 to 10 min using carbon dioxide.
Immunofluorescence
Tumor tissue-mounted slides were subjected to a graded ethanol series for permeabilization, involving sequential immersion in 100%, 90%, 80%, 70%, and 60% ethanol for 5 min each. Antigen retrieval was then performed using citrate buffer (pH 6.0) for 15–20 min via heat-induced epitope retrieval (HIER). Slides were subsequently cooled under running water and treated with 0.1% Triton X-100 for 10 min at room temperature to reduce non-specific background staining. After three washes in PBS (5 min each), endogenous peroxidase activity was blocked using Endoblocker (Peroxidase-Blocking Solution, cat. no. S2023; Dako, Glostrup, Denmark) for 20 min at room temperature. Slides were again washed with PBS (3 × 5 min), and then incubated overnight at 4 °C with the primary antibody against Ki-67 (cat. no. ab16667; Abcam, Cambridge, UK), diluted 1:300 in blocking buffer. The following day, after three additional PBS washes, the sections were incubated with the secondary antibody, anti-rabbit Alexa Fluor 568 (cat. no. A1101; Invitrogen, California, USA), diluted 1:200, for 1 h at room temperature. Nuclear counterstaining was performed using DAPI (1:500 dilution; cat. no. D1306; Life Technologies, California, USA) for 10 min. After a final PBS wash (3 × 5 min), slides were mounted using Paramount Aqueous Mounting Medium (cat. no. S3025; Dako, Life Technologies, California, USA) and covered with a coverslip. Mounted slides were allowed to dry at room temperature for 24 h. Fluorescence images were acquired using the EVOS FL Imaging System (Invitrogen, California, USA). The number of Ki-67–positive cell per 100 tumor cells was used to determine the Ki-67 labeling proliferation index (%).
Statistical analysis
The significance of experimental differences was assessed using an unpaired Student’s t-test. Data are presented as mean ± standard error. All experimental assays were conducted independently in triplicate. Statistical analyses were conducted using SPSS version 21.0 (IBM Corp.). A p-value of < 0.05 was considered statistically significant.
Continue Reading
-

Julie Joubert captures the French Foreign Legion’s new recruits
“Patria Nostra”—which means “our homeland”—continuously subverts the stereotype of a soldier and the hypermasculinity of militarized bodies with quiet moments of grace and intimacy. Her camera shifts between group scenes of drills to…
Continue Reading
-

Cheese Cave Fungi Help Scientists See Evolution In Real Time
What do you get when you propose to someone at a cheesemaking facility? Hopefully a yes, if they’re not too put off by the smell – but as a new study has shown, you might also end up with the ability to witness evolution in real time.
That’s…
Continue Reading
-

Double gold for Spain at the 2025 World Championships in 49er and 49er FX while Brits claim Nacra 17 title
Husband and wife team, Gimson and Burnet gain another gold band
For their part, Barceló and Cantero had the best possible start in their first competition as a team. The duo only began training this year with a view to the LA28 Olympics after…
Continue Reading
-

Breast Cancer Polygenic Risk Scores Awareness Lags – European Medical Journal Breast Cancer Polygenic Risk Scores Awareness Lags
HIGH-RISK women had limited awareness yet strong support for breast cancer polygenic risk scores in clinical risk assessment more.
Study Snapshot
In a survey of 828 women at elevated breast cancer risk without a prior diagnosis, only 18.5% had…
Continue Reading
-
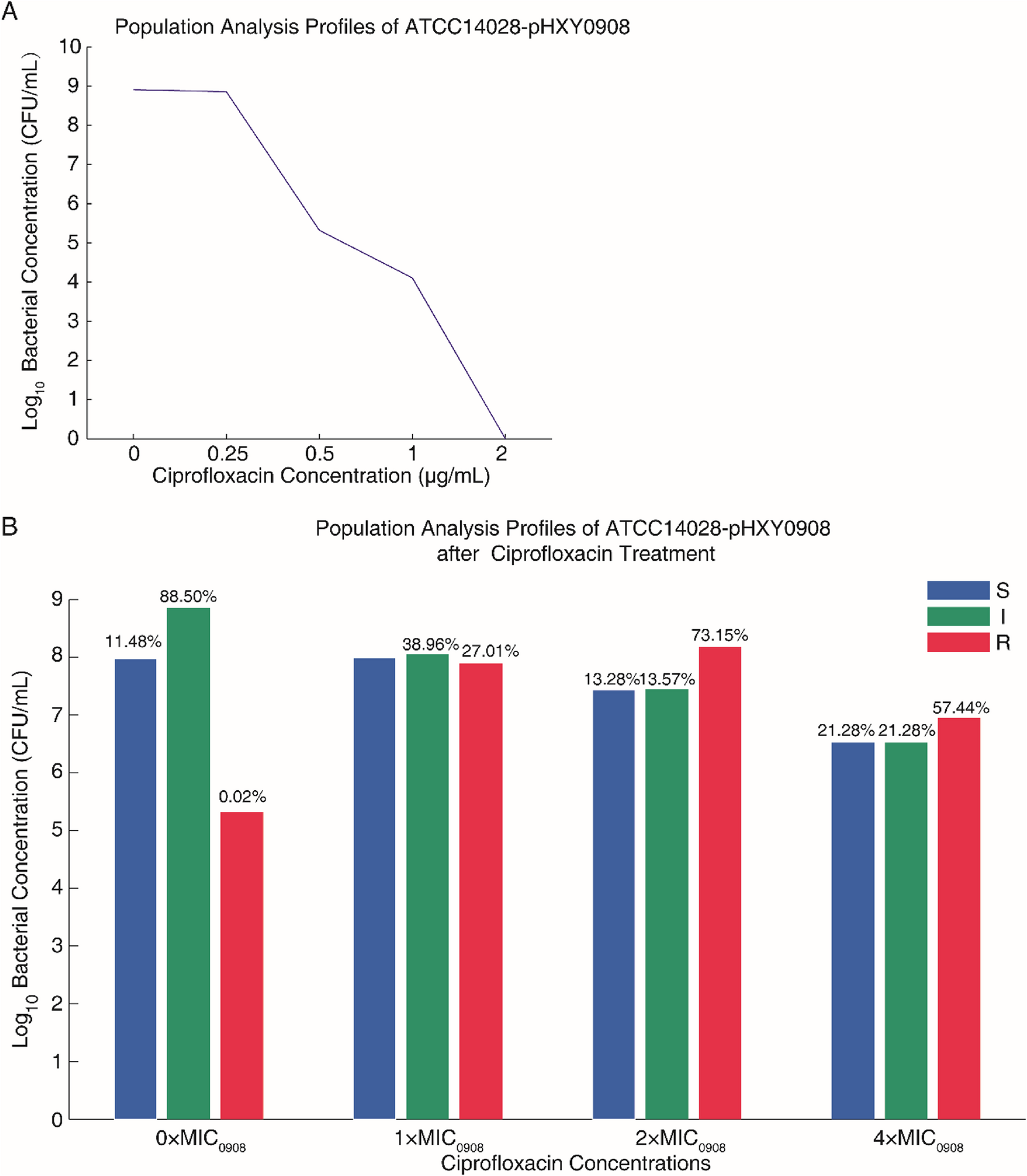
Plasmid pHXY0908 confers ciprofloxacin heteroresistance to Salmonella enterica serovar typhimurium ATCC 14028 by regulating efflux pump gene expression | BMC Microbiology
Chattaway MA, Gentle A, Nair S, Tingley L, Day M, Mohamed I, Jenkins C, Godbole G. Phylogenomics and antimicrobial resistance of Salmonella Typhi and paratyphi A, B and C in england, 2016–2019. Microb Genom 2021, 7(8):000633.
Xiang Y, Zhu K, Min K, Zhang Y, Liu J, Liu K, et al. Characterization of a Salmonella enterica serovar typhimurium lineage with rough colony morphology and multidrug resistance. Nat Commun. 2024. https://doi.org/10.1038/s41467-024-50331-y.
Google Scholar
Coburn B, Grassl GA, Finlay BB. Salmonella, the host and disease: a brief review. Immunol Cell Biol. 2007;85(2):112–8.
Google Scholar
Shi H, Zhou X, Zou W, Wang Y, Lei C, Xiang R, et al. Co-occurrence of biofilm formation and quinolone resistance in Salmonella enterica serotype typhimurium carrying an IncHI2-type oqxAB-positive plasmid. Microb Pathog. 2018;123:68–73.
Google Scholar
Chang MX, Zhang JF, Sun YH, Li RS, Lin XL, Yang L, et al. Contribution of different mechanisms to ciprofloxacin resistance in Salmonella spp. Front Microbiol. 2021;12:663731.
Google Scholar
Nambiar RB, Elbediwi M, Ed-Dra A, Wu B, Yue M. Epidemiology and antimicrobial resistance of Salmonella serovars typhimurium and 4,[5],12:i- recovered from hospitalized patients in China. Microbiol Res. 2024;282:127631.
Google Scholar
Kuang D, Zhang J, Xu X, Shi W, Chen S, Yang X, et al. Emerging high-level Ciprofloxacin resistance and molecular basis of resistance in Salmonella enterica from humans, food and animals. Int J Food Microbiol. 2018;280:1–9.
Google Scholar
Yu F, Chen Q, Yu X, Pan J, Li Q, Yang L, et al. High prevalence of plasmid-mediated quinolone resistance determinant aac(6’)-Ib-cr amongst Salmonella enterica serotype typhimurium isolates from hospitalised paediatric patients with diarrhoea in China. Int J Antimicrob Agents. 2011;37(2):152–5.
Google Scholar
Campillo R, Garcia-Penas I, Lopez N, Sanchez A, Fau A, Gomez D, et al. Ciprofloxacin-resistant Salmonella typhimurium demonstrates cross-tolerance to heat treatments in liquid food matrices. Food Res Int. 2025;210:116330.
Google Scholar
Andersson DI, Nicoloff H, Hjort K. Mechanisms and clinical relevance of bacterial heteroresistance. Nat Rev Microbiol. 2019;17(8):479–96.
Google Scholar
Lin JY, Zhu ZC, Zhu J, Chen L, Du H. Antibiotic heteroresistance in Klebsiella pneumoniae: definition, detection methods, mechanisms, and combination therapy. Microbiol Res. 2024;283:127701.
Google Scholar
El-Halfawy OM, Valvano MA. Antimicrobial heteroresistance: an emerging field in need of clarity. Clin Microbiol Rev. 2015;28(1):191–207.
Google Scholar
Chen Y, Hu D, Zhang Q, Liao XP, Liu YH, Sun J. Efflux pump overexpression contributes to Tigecycline heteroresistance in Salmonella enterica serovar typhimurium. Front Cell Infect Microbiol. 2017;7:37.
Google Scholar
Hjort K, Nicoloff H, Andersson DI. Unstable tandem gene amplification generates heteroresistance (variation in resistance within a population) to colistin in Salmonella enterica. Mol Microbiol. 2016;102(2):274–89.
Google Scholar
Weston N, Sharma P, Ricci V, Piddock LJV. Regulation of the AcrAB-TolC efflux pump in Enterobacteriaceae. Res Microbiol. 2018;169(7–8):425–31.
Google Scholar
Hernando-Amado S, Laborda P, La Rosa R, Molin S, Johansen HK, Martinez JL. Ciprofloxacin resistance rapidly declines in NfxB defective clinical strains of Pseudomonas aeruginosa. Nat Commun. 2025;16(1):4992.
Google Scholar
Suwanthada P, Kongsoi S, Jayaweera S, Akapelwa ML, Thapa J, Nakajima C, et al. Interplay between amino acid substitution in GyrA and QnrB19: elevating fluoroquinolone resistance in Salmonella typhimurium. ACS Infect Dis. 2024;10(8):2785–94.
Google Scholar
Strahilevitz J, Jacoby GA, Hooper DC, Robicsek A. Plasmid-mediated quinolone resistance: a multifaceted threat. Clin Microbiol Rev. 2009;22(4):664–89.
Google Scholar
Wong MH, Chan EW, Xie L, Li R, Chen S. IncHI2 plasmids are the key vectors responsible for OqxAB transmission among Salmonella species. Antimicrob Agents Chemother. 2016;60(11):6911–5.
Google Scholar
Wan Y, Myall AC, Boonyasiri A, Bolt F, Ledda A, Mookerjee S, Weisse AY, Getino M, Turton JF, Abbas H, et al. Integrated analysis of patient networks and plasmid genomes to investigate a regional, multispecies outbreak of Carbapenemase-Producing enterobacterales carrying both blaIMP and mcr-9 genes. J Infect Dis. 2024;230(1):e159–70.
Google Scholar
Treilles M, Chatre P, Drapeau A, Madec JY, Haenni M. Spread of the mcr-1 colistin-resistance gene in Escherichia coli through plasmid transmission and chromosomal transposition in French goats. Front Microbiol. 2022;13:1023403.
Google Scholar
Tan W, Lu Y, Zhu Z, Xu Z, Zhang Y, Huang Q, Meng X, Li S. Cotransfer of resistance to cephalosporins, colistin, and fosfomycin mediated by an IncHI2/pSH16G4928-like plasmid in ESBL-producing monophasic Salmonella typhimurium strains of pig origin. J Appl Microbiol 2023, 134(3):lxac060.
Deng L, Lv LC, Tu J, Yue C, Bai Y, He X, et al. Clonal spread of blaNDM-1-carrying Salmonella enterica serovar typhimurium clone ST34 and wide spread of IncHI2/ST3-blaNDM-5 plasmid in China. J Antimicrob Chemother. 2024;79(8):1900–9.
Google Scholar
Zhang CZ, Zhang Y, Ding XM, Lin XL, Lian XL, Trampari E, et al. Emergence of ciprofloxacin heteroresistance in foodborne Salmonella enterica serovar agona. J Antimicrob Chemother. 2020. https://doi.org/10.1093/jac/dkaa288.
Google Scholar
Zwe YH, Chin SF, Kohli GS, Aung KT, Yang L, Yuk HG. Whole genome sequencing (WGS) fails to detect antimicrobial resistance (AMR) from heteroresistant subpopulation of Salmonella enterica. Food Microbiol. 2020;91:103530.
Google Scholar
Langfelder P, Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics. 2008;9:559.
Google Scholar
Zhang B, Horvath S. A general framework for weighted gene co-expression network analysis. Stat Appl Genet Mol Biol. 2005;4:Article17.
Google Scholar
Zhang H, Ma S, Wang Y, Chen X, Li Y, Wang M, Xu Y. Development of an obesity-related multi-gene prognostic model incorporating clinical characteristics in luminal breast cancer. iScience. 2024;27(3):109133.
Google Scholar
Lieberman-Aiden E, van Berkum NL, Williams L, Imakaev M, Ragoczy T, Telling A, Amit I, Lajoie BR, Sabo PJ, Dorschner MO, et al. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science. 2009;326(5950):289–93.
Google Scholar
Lioy VS, Cournac A, Marbouty M, Duigou S, Mozziconacci J, Espeli O, et al. Multiscale structuring of the E. coli chromosome by nucleoid-associated and condensin proteins. Cell. 2018;172(4):771-e783718.
Google Scholar
Zhao X, Drlica K. Reactive oxygen species and the bacterial response to lethal stress. Curr Opin Microbiol. 2014;21:1–6.
Google Scholar
Hall CW, Zhang L, Mah TF. PA3225 is a transcriptional repressor of antibiotic resistance mechanisms in Pseudomonas aeruginosa. Antimicrob Agents Chemother. 2017. https://doi.org/10.1128/AAC.02114-16.
Google Scholar
Dewachter L, Fauvart M, Michiels J. Bacterial heterogeneity and antibiotic survival: understanding and combatting persistence and heteroresistance. Mol Cell. 2019;76(2):255–67.
Google Scholar
Lian X, Wang X, Liu X, Xia J, Fang L, Sun J, et al. oqxAB-positive IncHI2 plasmid pHXY0908 increase Salmonella enterica serotype typhimurium strains tolerance to ciprofloxacin. Front Cell Infect Microbiol. 2019;9:242.
Google Scholar
Calhoun LN, Kwon YM. Structure, function and regulation of the DNA-binding protein Dps and its role in acid and oxidative stress resistance in Escherichia coli: a review. J Appl Microbiol. 2011;110(2):375–86.
Google Scholar
Oh TJ, Jung IL, Kim IG. The Escherichia coli SOS gene SbmC is regulated by H-NS and RpoS during the SOS induction and stationary growth phase. Biochem Biophys Res Commun. 2001;288(4):1052–8.
Google Scholar
Labrou M, Michail G, Ntokou E, Pittaras TE, Pournaras S, Tsakris A. Activity of oxacillin versus that of vancomycin against oxacillin-susceptible mecA-positive Staphylococcus aureus clinical isolates evaluated by population analyses, time-kill assays, and a murine thigh infection model. Antimicrob Agents Chemother. 2012;56(6):3388–91.
Google Scholar
Wu X, Xu L, Gu W, Xu Q, He QY, Sun X, Zhang G. Iterative genome correction largely improves proteomic analysis of nonmodel organisms. J Proteome Res. 2014;13(6):2724–34.
Google Scholar
Zhang G, Fedyunin I, Kirchner S, Xiao C, Valleriani A, Ignatova Z. FansE: an accurate algorithm for quantitative mapping of large scale sequencing reads. Nucleic Acids Res. 2012;40(11):e83.
Google Scholar
Xiao CL, Mai ZB, Lian XL, Zhong JY, Jin JJ, He QY, et al. FANSe2: a robust and cost-efficient alignment tool for quantitative next-generation sequencing applications. PLoS ONE. 2014;9(4):e94250.
Google Scholar
Yang L, Lian X, Zhang W, Guo J, Wang Q, Li Y, Chen Y, Yin X, Yang P, Lan F, et al. Finding missing proteins from the epigenetically manipulated human cell with stringent quality criteria. J Proteome Res. 2015;14(9):3645–57.
Google Scholar
Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. Mapping and quantifying mammalian transcriptomes by RNA-seq. Nat Methods. 2008;5(7):621–8.
Google Scholar
Bloom JS, Khan Z, Kruglyak L, Singh M, Caudy AA. Measuring differential gene expression by short read sequencing: quantitative comparison to 2-channel gene expression microarrays. BMC Genomics. 2009;10:221.
Google Scholar
Zhao J, Zhang H, Qin B, Nikolay R, He QY, Spahn CMT, et al. Multifaceted stoichiometry control of bacterial operons revealed by deep proteome quantification. Front Genet. 2019;10:473.
Google Scholar
Robinson MD, McCarthy DJ, Smyth GK. EdgeR: a bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26(1):139–40.
Google Scholar
McCarthy DJ, Chen Y, Smyth GK. Differential expression analysis of multifactor RNA-seq experiments with respect to biological variation. Nucleic Acids Res. 2012;40(10):4288–97.
Google Scholar
Futschik ME, Carlisle B. Noise-robust soft clustering of gene expression time-course data. J Bioinform Comput Biol. 2005;3(4):965–88.
Google Scholar
Kumar L. Mfuzz: a software package for soft clustering of microarray data. Bioinformation. 2007;2(1):5–7.
Google Scholar
Ghazalpour A, Doss S, Zhang B, Wang S, Plaisier C, Castellanos R, Brozell A, Schadt EE, Drake TA, Lusis AJ, et al. Integrating genetic and network analysis to characterize genes related to mouse weight. PLoS Genet. 2006;2(8):e130.
Google Scholar
Mi H, Muruganujan A, Ebert D, Huang X, Thomas PD. PANTHER version 14: more genomes, a new PANTHER GO-slim and improvements in enrichment analysis tools. Nucleic Acids Res. 2019;47(D1):D419-26.
Google Scholar
Supek F, Bošnjak M, Škunca N, Šmuc T. Revigo summarizes and visualizes long lists of gene ontology terms. PLoS ONE. 2011;6(7):e21800.
Google Scholar
Ortet P, De Luca G, Whitworth DE, Barakat M. P2TF: a comprehensive resource for analysis of prokaryotic transcription factors. BMC Genomics. 2012;13:628.
Google Scholar
Continue Reading
