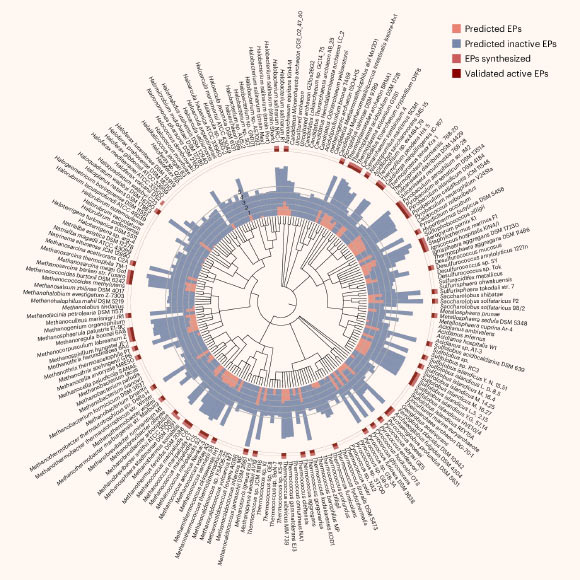
Antimicrobial resistance is one of the greatest threats facing humanity, making the need for new antibiotics more critical than ever. While most antibiotics originate from bacteria and fungi, Archaea offer a largely untapped reservoir for antibiotic discovery. In a new study, researchers at the University of Pennsylvania leveraged deep learning to systematically explore archaeal organisms; by mining proteomes of 233 archaeal species, they identified 12,623 molecules with potential antimicrobial activity.
Torres et al. synthesized 80 archaeasins, 93% of which showed antimicrobial activity in vitro against Acinetobacter baumannii, Escherichia coli, Klebsiella pneumoniae, Pseudomonas aeruginosa, Staphylococcus aureus and Enterococcus spp. Image credit: Torres et al., doi: 10.1038/s41564-025-02061-0.
“Previous efforts to find new antibiotics have looked mostly at fungi, bacteria and animals,” said Dr. César de la Fuente, a researcher at the University of Pennsylvania.
“In the past, we’ve used AI models to identify antibiotic candidates in a range of unlikely sources, from the DNA of extinct organisms to the chemicals in animal venom.”
“Now, we’re applying those tools to a new set of data: the proteins of hundreds of ancient microbes.”
“There’s a whole other domain of life waiting to be explored.”
Distinct from both bacteria and from eukaryotes (which include plants, animals and fungi), Archaea occupy their very own branch on the tree of life.
Though they resemble bacteria under a microscope, Archaea fundamentally differ in their genetics, cell membranes and biochemistry.
These differences allow them to survive in some of Earth’s most extreme environments, from superheated undersea vents to blistering hot springs like those in Yellowstone National Park.
Because Archaea often thrive where few other organisms can — enduring crushing pressures, toxic chemicals and extreme temperatures — their biology has evolved in unusual ways.
That makes them a promising but largely untapped source of new molecular tools, including compounds that may function like antibiotics but operate differently from those currently in use.
“We were drawn to Archaea because they’ve had to evolve biochemical defenses in unusual environments,” said Dr. Marcelo Torres, also from the University of Pennsylvania.
“We thought, if they’ve survived for billions of years under those conditions, maybe they’ve developed unique ways to fight off microbial competitors, and maybe we could learn from that.”
To uncover potential antibiotic compounds hidden in Archaea, the researchers turned to artificial intelligence.
They leveraged an updated version of APEX, an AI tool that they originally developed to identify antibiotic candidates in ancient biology, including in the proteins of extinct animals like the woolly mammoth.
Having seen thousands of peptides — short chains of amino acids — with known antimicrobial properties, APEX can predict the likelihood that a given sequence of amino acids will have similar effects.
By retraining APEX 1.1 on thousands of additional peptides and information about bacteria that cause diseases in humans, the scientists prepared the tool to predict which peptides in Archaea might inhibit bacterial growth.
Scanning 233 archaeal species yielded more than 12,000 antibiotic candidates.
The authors dubbed these molecules archaeasins, which chemical analysis revealed differ from known antimicrobial peptides (AMPs), in particular in their distribution of electric charge.
They then selected 80 archaeasins to test against actual bacteria.
“Trying to find new antibiotics one molecule at a time is like looking for needles in a haystack,” says Fangping Wan, a postdoctoral researcher at the University of Pennsylvania.
“AI speeds up the process by identifying where the needles are likely to be.”
Antibiotics work in a number of ways. Some punch holes in bacterial membranes, while others shut down the organisms’ ability to make proteins.
The researchers found that, unlike most known AMPs, which attack a bacterium’s outer defenses, archaeasins seem to pull the plug from the inside, scrambling the electrical signals that keep the cell alive.
In tests against a range of disease-causing, drug-resistant bacteria, 93% of the 80 archaeasins surveyed demonstrated antimicrobial activity against at least one bacterium.
The team then selected three archaeasins to test in animal models.
Four days after a single dose, the archaeasins all arrested the spread of a drug-resistant bacterium often acquired in hospitals.
One of the three compounds demonstrated activity comparable to polymyxin B, an antibiotic commonly used as a last-line of defense against drug-resistant infections.
“This research shows that there are potentially many antibiotics waiting to be discovered in Archaea,” Dr. de La Fuente said.
“With more and more bacteria developing resistance to existing antibiotics, it’s critical to find new antibiotics in unconventional places to replace them.”
A paper on the results was published today in the journal Nature Microbiology.
_____
M.D.T. Torres et al. Deep learning reveals antibiotics in the archaeal proteome. Nat Microbiol, published online August 12, 2025; doi: 10.1038/s41564-025-02061-0