Introduction – The Unfinished Story of RNA
In a recent lecture on RNA Biology by Dr. Fazal Adnan, the Head of the Department at Atta Ur Rahman School of Applied Biosciences in Islamabad, presented an intriguing illustration (see Figure 1), that truly ignited curiosity among all of us students. This captivating image not only piqued scientific curiosity but also inspired a deeper conversation about human molecular biology and its evolution. It made me reflect on both the incredible advancements we have made in the past and appreciate the journey of discovery that has brought us to where we are today.
It is a widely accepted practice to express gratitude to our ancestors for their contributions in advancing civilization and transforming our way of life. But what if I tell you that our bodies have a similar practice. Just like we honor those who came before us, our cells remember past infections through integrating invader sequences in our genome, helping our cells recognize and combat similar threats in the future. Isn’t that incredible? It’s a fascinating dance of human evolution alongside the evolution of microbes. So, who’s the smarter one in this partnership? Well, this is about the untold story of RNA biology that has revolutionized our understanding of molecular defense. Let’s dive into it.
A recent article published in Nature raised a critical question: Why is RNA structure so difficult to predict? While many bioinformatics platforms have successfully tackled protein analysis, they fall short in RNA prediction. This discrepancy highlights a significant challenge in our field, and we must delve into this question, as understanding RNA structure is key to advancing our knowledge in molecular biology and therapeutics.
For many years, RNA has served as an intermediary component within the framework of the central dogma of molecular biology. Then the discovery of ribozymes, the identification of RNA as a catalyst, and the understanding of the RNA world hypothesis have significantly heightened scholarly interest in the study of RNA.
The true importance of RNA began to shine through in 1974, when groundbreaking research unveiled its remarkable ability to self-modify. This unique trait allows RNA to carry out crucial functions that help safeguard the integrity of the cell and its entire cellular system. This process of regulation through inducing chemically modified tags annotates the sequence to functionalize and generate a desired product. The Accumulation of sequences from past generations has aided in the evolution of the genome to increase its fitness. This fascinating interplay reveals just how adaptable and vital RNA is to life itself.
This has led to the emergence of Epitrancriptomics, which encompasses post-transcriptional modifications that do not affect the base sequence. These modifications have significant implications for the final structure, stability, and translation efficiency of RNA molecules. These chemical modifications control gene expression beyond the DNA sequence.
Cracking The RNA Code: What Is Epitranscriptomics?
The genetic code can be likened to a language that narrates the experiences and resilience of human beings throughout their lives (see Figure 2). This code, composed of the four nucleotides adenine (A), uracil (U), guanine (G), and cytosine (C), conveys essential biological information. Furthermore, RNA modifications function as grammatical rules that enhance and clarify the meanings conveyed by the genetic sequence. To date, over 170 distinct types of RNA modifications have been identified across various forms of RNA, underscoring the intricate nature of genetic regulation. These modifications are not just static; they’re dynamic and reversible, tightly orchestrated by specialized enzymes. You can think of them as the storytellers in this biological tale: where “writers“(METTL3, METTL14) introduce transformative edits, “erasers“ (FTO, ALKBH5) refine them, and “readers” (YTHDF1, YTHDF2, IGF2BP1) interpret their significance. Embracing this complexity opens up a deeper understanding of the full spectrum of genetic expression.
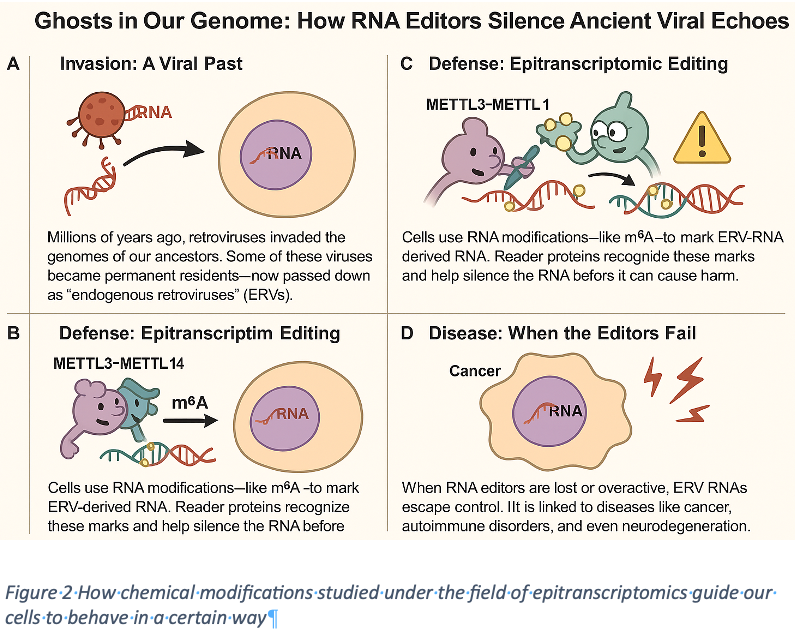
Epitranscriptomics is an intriguing RNA editing mechanism that sets itself apart from epigenetics. While epigenetics focuses on how chemical modifications influence gene expression, epitranscriptomics delves into the realm of RNA itself. It’s a fascinating layer of gene regulation that actively transforms the RNA message, shaping its destiny in unique ways. Imagine the power of writing directly on RNA, altering its fate and function; this is the dynamic world of epitranscriptomics!
Many different chemical changes can happen to RNA, but N6-methyladenosine stands out as a very important molecular switch. This modification plays a critical role, allowing the cell to adapt to its dynamic needs.
On the other hand, alterations like acetylation and pseudouridylation improve RNA’s regulatory capabilities even more. They increase the stability of the molecule, enhance its ability to fold, and maintain the integrity of its intricate two- and three-dimensional structures. Because of its crucial function in post-transcriptional control, this vast panorama of alterations highlights the immense complexity and distinctiveness of RNA. A relevant question is raised: might these RNA alterations play a significant role in clarifying the mechanisms behind aging, disease, and evolutionary processes? A pertinent inquiry arises: could these RNA modifications serve as key factors in elucidating the mechanisms underlying disease, aging, and evolutionary processes?
The Groundbreaking Discoveries That Changed RNA Biology
The epitranscriptomics and its application in research and development have opened new avenues and have answered some of the important questions on evolution and the role of molecular agents in it. The most significant application is the development of technology to map all the chemical modifications on the transcriptome, which can tell a lot about human health and disease and the adaptive trails a cell undergoes. For example, if we want to study cancer, we can check for RNA alterations, to seek specific modifications specific to that pathogenesis could help to develop novel targets for drug development and understanding how a cell undergoes dysfunction.
One of the most significant and potent examples is the development of the COVID-19 vaccine, where chemical alterations have been made to mRNA coding for the protein of interest, increasing the vaccine efficacy while minimizing the unwanted immune reactions. This is the way that understanding chemical modifications for safety profiles of many potent vaccines and therapeutic drugs accelerates the research advancement and provides a novel platform to tackle critical health concerns.
The Living and the Dead Li’s Elements
Historically, the epitranscriptomic encompasses the chemical changes such as methylation, acetylation, and pseudouridinylation, but now it has added some new modes of changes. Xiong et al reported that this region also includes RNA control by retrotransposition, which is assisted by the m6A alteration. These retrotransposons provide prospective targets for RNA modification.
A study by Dominissini et al distinguishes between “living” (retrotranspositionally competent) and “dead” (retrotranspositionally incompetent) L1 components where data indicates that m6A enhances the activity of functional “living” L1 sequences while simultaneously serving a significant role for “dead” L1 sequences. These “dead” L1 sequences can influence the host organism by inhibiting genes that typically suppress the transposition of “living” L1s. Moreover, these alterations exhibit non-random characteristics and are preserved, signifying that their involvement in retrotransposition represents a strategic framework designed to promote genomic stability and foster functional diversity.
VIP pass to tiny RNA molecules
Small RNAs, known for their existence in plants and traditional therapeutic practices, face considerable obstacles when they travel through the human digestive system. These small compounds are naturally delicate and have difficulty passing through the membranes, which causes their degradation through the body’s surveillance system, before they can induce their potential effects. Despite facing numerous challenges, the remarkable potential of these biomolecules to transform various aspects is truly promising and should not be overlooked.
Guo et al performed research on Ban Zhi Liana, a herb in traditional Chinese medicine where the isolated crude extract contains the specifically modified small RNAs. These are natural, particularly the two fascinating types: 2′-O-methylation (2′-O-Me) and N6-methyladenosine (m6A). Furthermore, the investigation has also found an additional modification known as 5-methylcytidine (m5C); however, this particular modification did not demonstrate the same exceptional advantages as the others. These findings offer a promising perspective on the potential of natural compounds to advance health and therapeutic applications.
The Next Frontier: Can We Hack RNA Modifications for Medicine?
In the above mentioned context, the field of epitranscriptomics is rapidly evolving, providing great opportunities for revealing previously unknown facts within scientific inquiry. Looking ahead, we may anticipate the development of novel instruments and assays, which will pave the way for further in-depth study in RNA biology, covering topics such as aging and cancer, allowing the detection of changes at the individual cell level. Such breakthroughs will allow us to monitor organ health with great accuracy. The advancement of knowledge and technologies to map all chemical changes in a transcriptome will enable the screening of lethal and healthy tags, and techniques to restore them. Here, CRISPR-based tools are transforming the utilization of chemical tags from diagnostics to therapeutics, allowing researchers to add or remove critical modifications with precision. This advancement is laying the groundwork for next-generation theragnostics, in which physicians may one day be able to address diseases by directly targeting RNA and making diagnostics more sensitive and precise.
We are just beginning to unlock the mysteries of RNA and its critical role in understanding how our bodies function. In the not-so-distant future, we will gain insights into how our bodies heal, all thanks to the fascinating world of RNA biology. It’s an exciting time to be part of this journey.
References
Bhat, S. S., Bielewicz, D., Jarmolowski, A., & Szweykowska-Kulinska, Z. (2018). N6-methyladenosine (m6A): Revisiting the Old with Focus on New, an Arabidopsis thaliana Centered Review. Genes, 9(12), 596. https://doi.org/10.3390/genes9120596
Cerneckis, J., Ming, G.-L., Song, H., He, C., & Shi, Y. (2024). The rise of epitranscriptomics: Recent developments and future directions. Trends in Pharmacological Sciences, 45(1), 24–38. https://www.cell.com/trends/pharmacological-sciences/fulltext/S0165-6147(23)00254-7
Dominissini, D., Moshitch-Moshkovitz, S., Schwartz, S., Salmon-Divon, M., Ungar, L., Osenberg, S., Cesarkas, K., Jacob-Hirsch, J., Amariglio, N., & Kupiec, M. (2012). Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature, 485(7397), 201–206. https://idp.nature.com/authorize/casa?redirect_uri=https://www.nature.com/articles/nature11112&casa_token=998Qh-5Va2cAAAAA:InO38ZnffREIuw_ScTtRKxGpwzFdbabgd0mQRmMDeDoJIkshEt5L39MG4Lc2K_sXGYe8FtxUM293sYqm0g
Guo, S., Li, Z., Li, X., Liang, Z., Zhao, D., Sun, N., Liu, J., Wang, X., Mei, S., & Qiao, X. (2025). 2′-O-methylation and N6-methyladenosine enhance the oral delivery of small RNAs in mice. Molecular Therapy Nucleic Acids. https://www.cell.com/molecular-therapy-family/nucleic-acids/fulltext/S2162-2531(25)00128-3
Kwon, D. (2025). RNA function follows form-why is it so hard to predict? Nature, 639(8056), 1106–1108. https://pubmed.ncbi.nlm.nih.gov/40128371/
Xiong, F., Wang, R., Lee, J.-H., Li, S., Chen, S.-F., Liao, Z., Hasani, L. A., Nguyen, P. T., Zhu, X., & Krakowiak, J. (2021). RNA m6A modification orchestrates a LINE-1–host interaction that facilitates retrotransposition and contributes to long gene vulnerability. Cell Research, 31(8), 861–885. https://www.nature.com/articles/s41422-021-00515-8