A deep-learning algorithm unravels a collection of archaeasins, peptides from the archaeal proteome with potential antimicrobial activity and implications for the development of next-generation antibiotics
You have full access to this article via your institution.
Antimicrobial resistance (AMR) is an emerging threat for global health, as it is associated with up to four million deaths each year1. One of the greatest challenges in combating AMR is the rise of multidrug-resistant bacteria, which are exhausting the options for treating bacterial infections. Therefore, the discovery of new antibiotics is critical. Unfortunately, antibiotic discovery and development have notably slowed in recent decades owing to scientific, technical and financial barriers2. As a result, there is an urgent need for innovative, cost-effective strategies to uncover novel antimicrobial agents.
A promising approach is the exploration of encrypted peptides (EPs) as potential antibiotics3. EPs are bioactive sequences embedded in larger precursor proteins. They do not exhibit activity in their native form but can be released by enzymatic cleavage. Once liberated, these peptides may exhibit various biological activities, including antimicrobial effects. The properties of these peptides have been investigated for EPs derived from bovine proteins, especially4; for instance, lactoferricin is generated from the protein lactoferrin and exhibits strong antimicrobial activity5. Despite their potential, no EP has been approved for use as a systemic antibiotic yet, although several candidates are under investigation.
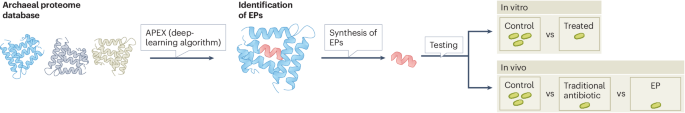
In this issue of Nature Microbiology, Torres et al.6 present a new collection of candidate antimicrobial EPs. The authors used an updated version of their deep-learning framework, APEX, to identify thousands of putative antimicrobial peptides (Fig. 1). A previous version of APEX had already enabled this research group to discover antimicrobial peptides in the proteomes of extinct organisms7. The new version of APEX predicts bacterial strain-specific antimicrobial activity for peptides based on previously published datasets and provides potential minimum inhibitory concentration values. The novelty of this study lies in the source: a dataset of 233 archaeal proteomes derived from curated public databases. Unlike bacteria and fungi, archaea have been largely overlooked as potential sources of antibiotics. Yet, their unique structural and metabolic features suggest that archaeal-derived compounds may offer entirely new classes of antimicrobials, which could be vital in the fight against AMR.
Using a curated database of archaeal proteomes, a collection of EPs has been predicted using APEX, a deep-learning framework. EPs are embedded in larger proteins, and once they are released can have different bioactive properties. After the identification of potential antimicrobial EPs, a selection was tested with in vitro and in vivo models, showing antimicrobial activity.
Using this dataset, Torres et al. identified more than 12,000 putative antimicrobial peptides, which they named archaeasins. Of these, 80 were selected for further testing based on their antimicrobial potential (based on the minimum inhibitory concentration values) and sequence diversity compared with previously known antimicrobial peptides. In vitro assays revealed that nearly all tested archaeasins exhibited antimicrobial activity against at least one clinically relevant pathogen, such as Acinetobacter baumannii, Escherichia coli, Klebsiella pneumoniae, Pseudomonas aeruginosa, Staphylococcus aureus, Enterococcus faecalis or Enterococcus faecium. Notably, the peptides also demonstrated low toxicity towards two human cell lines — red blood cells and embryonic kidney cells. Furthermore, the authors evaluated the efficacy of several archaeasins in preclinical animal models (a skin abscess and deep thigh infection model) infected with A. baumannii. Archaeasin-73 showed particularly promising results, with antimicrobial effects higher than levofloxacin and comparable to those of polymyxin B, two different antibiotics used as positive controls for those animal infection models.
In spite of the promising results, challenges remain for translating EPs into viable therapeutics. The candidate molecules must fulfil several requirements, such as low toxicity, stability, optimal delivery to the organism and effectiveness. For that, more research is needed regarding the pharmacokinetics and metabolic behaviour of each EP. Another important aspect is the potential emergence of resistance to the new molecules. Nevertheless, this research opens exciting new possibilities in the ongoing battle against AMR. Indeed, scientists are testing different kinds of molecules, including other peptides, to try to discover previously unknown classes of antibiotics. A promising example is the recent discovery of a lasso peptide — a class of peptides with threaded-loop topology similar to lassos — with activity against A. baumannii8.
A key insight from the study by Torres et al. is the importance of environmental microorganisms — especially thermophilic archaea — as potential sources of new bioactive compounds. The authors observed that some thermophilic archaea encode a higher proportion of EPs with predicted antimicrobial activity. This finding underscores the key role that environmental microbiology can play in laying the groundwork for future biotechnological breakthroughs. This is not a new idea; for example, in the 1960s, Brock and Freeze isolated Thermus aquaticus from a Yellowstone geyser9. Decades later, the heat-stable polymerase from this organism became a cornerstone of polymerase chain reaction (PCR) — a technology now indispensable across biomedical and forensic sciences.
However, one limitation for new discoveries is the dependence on cultured organisms. As the authors note, their analysis was restricted to curated proteomes from cultured archaea, thereby excluding vast groups of uncultured microorganisms, including complete phyla. According to the ‘great plate anomaly’ paradigm, fewer than 1% of microbial species are currently culturable10. Culturing efforts are improving and continued work in this space could help unlock an enormous reservoir of unknown bioactive molecules.
In sum, the work by Torres et al. represents a promising step in the search for new antibiotics. Their study not only demonstrates the utility of their computational framework for antimicrobial peptide discovery but also expands the scope of potential sources to include previously overlooked domains of life.