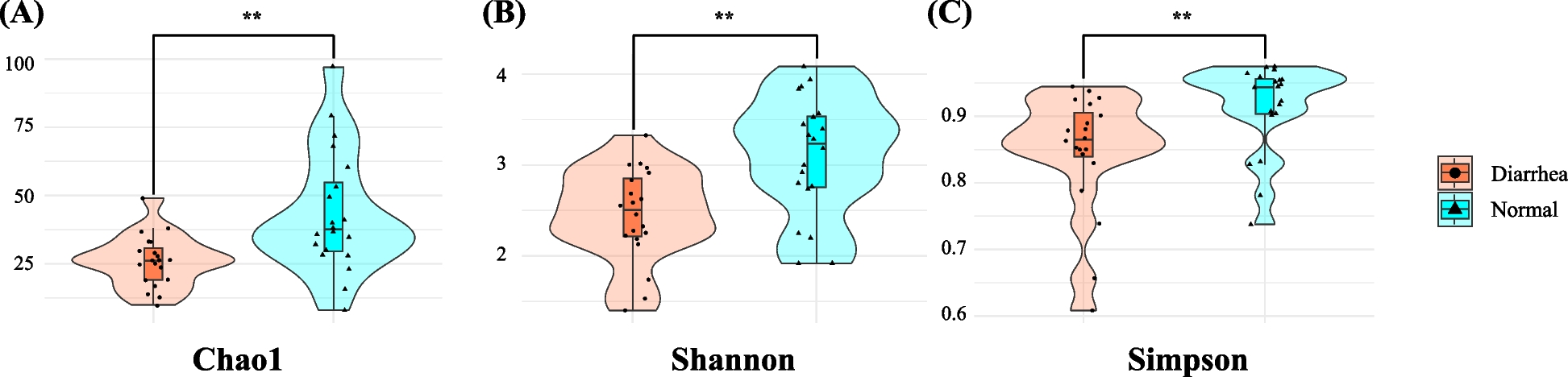
In this study, we investigated the potential association of NCD with shifts in the fecal microbiota and characterized these microbial changes at the taxonomic and functional levels. Our analysis of alpha diversity revealed a significantly reduced fecal microbiota diversity in diarrheic calves than in normal calves. The diarrheic group showed lower richness (Chao1) and evenness (Shannon and Simpson indices), indicating an association between NCD and significantly reduced microbial diversity. Lower alpha diversity often signifies a dysbiotic state, implying disrupted gut microbiota balance [23]. This may be related to the overgrowth of pathogenic or opportunistic bacteria and the concurrent depletion of beneficial or commensal taxa [23]. Such disruptions in early life may be associated with alterations in gut barrier function, nutrient utilization, and immune regulation, which could be linked to diarrheal episodes [23].
Our findings align with those in earlier reports demonstrating the correlation between NCD and diminished microbial diversity [11]. Underscoring the importance of a robust and diverse microbiota for gut health in calves. Reduced alpha diversity may serve as an indicator of gut health and disease susceptibility in neonatal calves [3]. Practically, monitoring these diversity indices may help identify at-risk animals sooner, facilitating timely interventions [24]. Additionally, strategies to preserve or restore microbial richness, such as the use of selective probiotics, prebiotics, and optimized feeding practices, may help mitigate diarrhea onset or severity and improve both calf welfare and farm productivity [25,26,27].
PCoA based on Bray–Curtis and Jensen–Shannon divergence measures indicated clear compositional differences in the fecal microbiota between normal and diarrheic calves. Despite some overlap, the two groups clustered distinctly, with diarrheic samples generally occupying more positive values on axis 1 and normal samples more negative values. The centroids for each group remained separate, indicating a marked shift in microbial community structure [11]. These clustering patterns suggest that NCD is associated with a broad restructuring of the gut microbial ecosystem, rather than merely a reduction or enrichment of a few specific taxa [2, 3]. In particular, the divergence along axis 1, which explained a substantial portion of the total variance, suggested the presence of group-specific microbial assemblages. Similar results in earlier studies on calf diarrhea [2, 3, 11] corroborate the notion of a distinct microbiota composition in diarrheic calves compared with their healthy peers, potentially reflecting pathogen overgrowth and beneficial microbe depletion. In practice, rapid microbiota profiling can help identify at-risk calves before clinical signs appear, enabling targeted management or therapeutic interventions [24]. Moreover, the pronounced shift in microbial community composition may inform future studies aimed at developing microbial or metabolite-based biomarkers for NCD.
Our results indicated that NCD is associated with disruptions in gut microbiota homeostasis, characterized by reduced abundances of beneficial taxa generally considered beneficial namely, those associated with gut health and anti-inflammatory effects (e.g., Faecalibacterium, Lawsonibacter) [11], and increased abundances of opportunistic or pathogenic bacteria (e.g., Escherichia, Salmonella, Klebsiella) [2, 3]. The LEfSe results further highlighted these differences, with E. coli and S. enterica exhibiting high LDA scores in diarrheic samples. These findings highlight association between NCD and disruptions in the gut microbiota, particularly an overrepresentation of opportunistic or pathogenic bacteria and a reduction in potentially beneficial taxa [3]. The increased prevalence of E. coli, S. enterica, and Klebsiella spp. aligns with their known roles in gastrointestinal diseases [2, 3], whereas the decline in Faecalibacterium and Lawsonibacter supports evidence that reduced beneficial microbes may impair gut barrier integrity and immune homeostasis [11]. Moreover, Lactobacillus and Limosilactobacillus, which are often considered beneficial under balanced conditions, were unexpectedly abundant in diarrheic calves, potentially due to small-intestinal mucosal damage leading to decreased lactase activity and the resultant increase in residual lactose in the colon [28, 29]. This elevated lactose could favor the proliferation of lactic acid bacteria under inflammatory or dysbiotic conditions [28, 29]. Furthermore, previous study has indicated that Lactobacillus abundance may increase during the recovery phase from diarrhea [3], suggesting that the higher abundance observed in our diarrheic calves might also reflect a compensatory or transitional response. Taken together, these findings indicate that the role of Lactobacillus in NCD is likely context-dependent, and further longitudinal studies are warranted to distinguish pathological overgrowth from recovery-associated expansion.
The LEfSe results corroborated these patterns, showing that specific taxa not only dominate in diseased states but may also exert an outsized influence on the overall microbial community structure. Pronounced shifts in bacterial composition underscore the importance of maintaining a balanced gut microbiota for neonatal calf health [30]. From an academic perspective, these results add to the growing body of evidence that NCD involves both established pathogens (e.g., Escherichia and Salmonella) and lesser-known contributors (e.g., Limosilactobacillus) [2, 11]. Practically, routine microbiota monitoring may enable the earlier identification of calves at risk and provide preliminary insights for potential interventions, such as probiotics or prebiotics, to restore beneficial taxa or suppress potential pathogens [31]. Moreover, the differential abundance of certain species, including E. coli, L. reuteri, S. enterica, and K. pneumoniae on the pathogenic side, and F. prausnitzii, and L. asaccharolyticus as potential beneficial indicators could inform future development of diagnostic panels or therapeutic interventions.
The results of the present study identified several metabolic pathways that differed significantly between diarrheic and normal calves (Fig. 5). Diarrheic samples displayed higher predicted abundances of pathways related to carbohydrate breakdown, specifically the Entner–Doudoroff pathway, hexitol fermentation, N-acetylneuraminate degradation, pentose phosphate pathway and dTDP–N–acetylhomosamine biosynthesis. These results suggest that the gut microbiota of diarrheic calves is functionally oriented toward rapid carbohydrate utilization and fermentation, possibly reflecting an inflammatory or dysbiotic state conducive to opportunistic microbes [3, 32]. The increased enrichment of the pathways, such as the Entner–Doudoroff pathway and hexitol fermentation, indicates a greater capacity to convert various sugars into energy and fermentation end products, potentially contributing to the clinical manifestations of diarrhea (e.g., increased osmotic pressure and altered short-chain fatty acid profiles) [33, 34]. The shifts in these functional pathways align with the compositional changes observed in diarrheic calves, particularly the overabundance of Escherichia and Salmonella, as many of these taxa thrive on simple carbohydrates such as those present under inflamed gut conditions [35,36,37]. Compared to alpha diversity, which reflect global community structure but offer limited insight, pathway-based profiling may provide more information for targeted intervention. Monitoring specific metabolic signatures may enable earlier detection of dysbiosis and better inform dietary or therapeutic strategies for NCD management. Understanding and monitoring these functionally enriched pathways may provide a basis for exploratory diagnostic tools and inform future intervention strategies to mitigate dysbiosis and its clinical consequences in NCD.
Nevertheless, our study also has some limitations. First, although our sample size (20 normal and 20 diarrheic calves) provided valuable insights, larger-scale investigations may offer greater statistical power to detect subtle microbial shifts. Second, our study did not incorporate longitudinal sampling; hence, we cannot definitively conclude how the microbiota evolves from pre-diarrheic to post-recovery stages. Third, although we performed functional predictions using PICRUSt2, which infers gene content from 16S rRNA profiles, these predictions do not measure actual gene expression or metabolic activity. Future studies incorporating higher-resolution approaches such as shotgun metagenomics or metatranscriptomics will be valuable to confirm the functional relevance of the microbial shifts observed and to explore gene-level and pathway-specific mechanisms involved in NCD pathogenesis. Therefore, interpretations regarding enriched metabolic pathways should be considered hypothetical and warrant confirmation though metagenomic, transcriptomic, or metabolomic approaches. Fourth, although diarrheic calves in this study were tested negative for major pathogens (bovine rotavirus, bovine coronavirus, Cryptosporidium, Giardia, and E. coli K99) using a rapid antigen detection kit, other infectious etiologies may not have been fully excluded. Therefore, the observed microbial shifts may have been partially influenced by undiagnosed enteric infections, and interpretation of causal relationships should be approached with caution. Future studies should employ longitudinal designs to track microbiota changes from birth through recovery and clarify the cause-and-effect relationships in NCD. Multi-omics approaches (e.g., shotgun metagenomics and metabolomics) can confirm the functional capacities suggested by predictive tools such as PICRUSt2. Fifth, extending this study to different geographical regions and farm management systems may enhance the generalizability of our findings and foster more robust NCD prevention strategies. Additionally, because the calves ranged in age from 2 days to 2 months, age-related physiological changes may have influenced the gut microbiota composition. However, due to the absence of data regarding precise day-of-age, a direct age-matching analysis between groups could not be conducted. While all calves were within the neonatal period, it is known that microbial communities undergo rapid maturation during this time. Therefore, we cannot exclude the possibility that some of the observed microbial variation was due to age-related dynamics rather than solely diarrheal status. Finally, fecal consistency was diagnosed by a veterinarian during routine examinations. However, a standardized scoring system was not applied, and additional health indicators (e.g., body temperature, appetite, mental state) were not systematically recorded. These limitations reduce the granularity of clinical assessments.