Species identification
The morphology and morphometric data of the present specimens of M. hemirhamphi are almost identical to the previous descriptions of this species [30, 53] regarding several features, including the size of trunk and proboscis receptacle, the shape (size) and armature of the proboscis, the number of transverse rows of trunk spines, the length of lemnisci, the shape and length of testis, the number of cement glands, the size of eggs, and the presence of caudal appendage in female (see Table 1 for details). Moreover, the present specimens were also collected from the type host of M. hemirhamphi, Hemiramphus intermedius (Cantor) (Beloniformes: Hemiramphidae). Therefore, we consider that the present specimens are conspecific with M. hemirhamphi. However, there has been no genetic data of M. hemirhamphi available on the GenBank database; thus, we could not further identify these specimens using the molecular-based method.
The morphology and morphometric data of the present specimens of R. laterospinosus agreed well with the previous descriptions of this species [10, 28, 31, 32] regarding several features, including the size of trunk and proboscis receptacle, the shape (size) and armature of the proboscis, the length of lemnisci, the shape and length of testis, the number of cement glands, and the size of eggs (see Table 2 for details). Additionally, comparison of the cox1 data of the present material and R. laterospinosus available in GenBank (MK572741-MK572744, LC777823, OR625530, and OR625531) showed a very high level of similarity (97.9–98.9%). Consequently, we considered our material belonging to R. laterospinosus.
Characterization of the complete mitogenomes of M. hemirhamphi and R. laterospinosus
The complete mitogenomes of M. hemirhamphi and R. laterospinosus both include 36 genes, containing 12 PCGs (cox1-3, nad1-6, nad4L, cytb, and atp6; missing atp8), 22 transfer RNA (tRNA) genes, and two ribosomal RNAs (rrnS and rrnL), plus two noncoding regions (SNCR is 160 bp, located between trnQ and trnY in M. hemirhamphi versus SNCR, which is 244 bp, between trnK and trnE in R. laterospinosus; LNCR is 4,238 bp, located between trnK and trnV in M. hemirhamphi versus LNCR, which is only 252 bp, located between trnQ and trnY in R. laterospinosus) (Fig. 2, Table 4). All mitochondrial genes are encoded on the same strand in the same direction. The lengths of the complete mitogenomes of M. hemirhamphi and R. laterospinosus are 17,272 bp and 13,567 bp, respectively. The overall A + T contents of M. hemirhamphi and R. laterospinosus mitogenomes are 60.5% and 62.9%, which both have a strong A + T bias. The nucleotide contents of the mitogenomes of M. hemirhamphi and R. laterospinosus are provided in Tables 4 and 5.
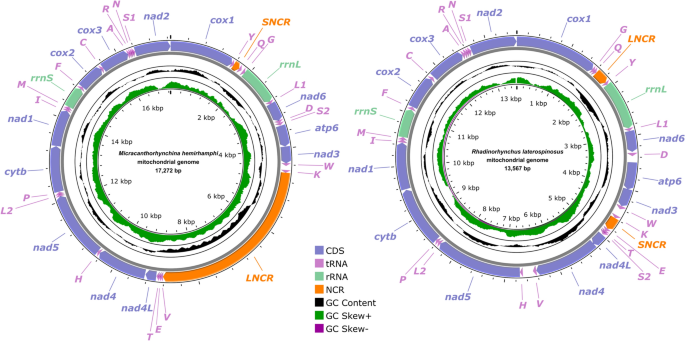
Gene map of mitochondrial genomes of Micracanthorhynchina hemirhamphi and Rhadinorhynchus laterospinosus. All 22 tRNA genes are nominated by the one-letter code with numbers differentiating each of the two tRNAs serine and leucine. All genes are transcribed in the clockwise direction on the same strand. The outermost circle shows the GC content, and the innermost circle shows the GC skew
The 12 PCGs in the mitogenomes of M. hemirhamphi and R. laterospinosus have 10,123 bp and 10,041 bp (excluding termination codons), and encode 3,372 and 3,345 amino acids, respectively. The size of each of the 12 PCGs in the mitogenomes of M. hemirhamphi and R. laterospinosus are provided in Table 5. Among the 12 PCGs of M. hemirhamphi, seven genes (cox1, cox2, cox3, nad2, nad4, nad6, and atp6) use GTG as the start codon, followed by two genes (cytb and nad5) that use ATA, while nad1, nad3, and nad4L use ATG, GTT, and TTG as the start codon, respectively. A total of four genes (cox1, cox3, nad2, and nad6) use TAA as the termination codon, and only atp6 uses TAG; the remaining seven genes (nad1, nad3, nad4, nad4L, nad5, cox2 and cytb) are inferred to terminate with incomplete stop codon T (Table 4). Among the 12 PCGs of R. laterospinosus, five genes (cox1, cox3, nad4, nad5, and nad6) use GTG as the start codon, followed by four genes (atp6, cytb, nad1, and cox2) that use ATG, while two genes (nad2 and nad4L) use TTG, and only nad3 uses ATT as the start codon. A total of four genes (cox1, nad5, nad6, and atp6) use TAG as the termination codon, followed by three genes (cox2, cox3, and nad4L) that use TAA, while only nad4 uses the incomplete stop codon TA. The remaining four genes (nad1, nad2, nad3, and cytb) are inferred to terminate with an incomplete stop codon T (Table 4). The codon usage in the mitogenomes of M. hemirhamphi and R. laterospinosus is presented in Fig. 3.
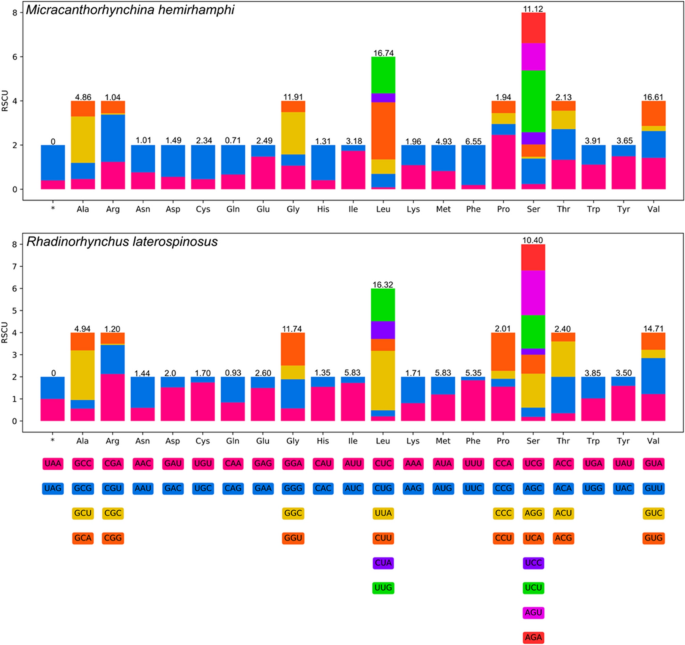
Relative synonymous codon usage (RSCU) of Micracanthorhynchina hemirhamphi and Rhadinorhynchus laterospinosus. The codon families (in alphabetical order) are labeled on the x-axis. Values on the top of each bar represent amino acid usage in percentage
The lengths of 22 tRNAs in the mitogenomes of M. hemirhamphi and R. laterospinosus are from 50 to 66 bases (Table 2). Their anticodon secondary structures are provided in Supplementary Material Figs. S1 and S2. In the mitogenomes of M. hemirhamphi and R. laterospinosus, the small ribosomal RNA gene (rrnS) is located between trnM and trnF in both species, with 514 bp to 536 bp in length, respectively; while the large ribosomal RNA gene (rrnL) is located in a different position with similar length in the two species (917 bp long, between trnG and trnL1 in M. hemirhamphi, versus 916 bp long, between trnY and trnL1 in R. laterospinosus).
In the mitogenomes of M. hemirhamphi and R. laterospinosus, the gene arrangements of 12 PCGs and two rRNAs are in the typical order of acanthocephalans: cox1, rrnL, nad6, atp6, nad3, nad4L, nad4, nad5, cytb, nad1, rrnS, cox2, cox3, and nad2. However, translocations of several tRNAs occurred in both mitogenomes of M. hemirhamphi and R. laterospinosus (Figs. 2 and 4).

Comparison of the linearized mitochondrial genome arrangement for acanthocephalans species (a total of 18 types of 36 gene arrangement in the mitogenomes of Acanthocephala reported before this study). All genes are transcribed in the same direction from left to right. The tRNAs are labelled by a single-letter code for the corresponding amino acid. Micracanthorhynchina hemirhamphi and Rhadinorhynchus laterospinosus are indicated using asterisk (*)
Phylogenetic analyses
The topologies of phylogenetic trees based on the amino acid sequences of 12PCGs using ML and BI methods, are nearly identical, which both support the division of the phylum Acanthocephala into three major monophyletic clades (clades I, II, and III) (Fig. 5). Clade I includes Macracanthorhynchus hirudinaceus, Oncicola luehei, Moniliformis tupaia, and Moniliformis sp., representing the class Archiacanthocephala. Clade II includes species of the orders Gyracanthocephala and Neoechinorhynchida, belonging to the class Eoacanthocephala, together with Polyacanthorhynchus caballeroi (a member of the class Polyacanthocephala). Clade III contains representatives of the orders Echinorhynchida and Polymorphida, representing the class Palaeacanthocephala. Micracanthorhynchina hemirhamphi and M. dakusuiensis clustered together, representing the family Micracanthorhynchinidae. The rhadinorhynchid species R. laterospinosus formed a sister relationship with a member of the Cavisomatidae (Cavisoma magnum) with maximum support.

Phylogenetic results using ML and BI methods based on the concatenating amino acid sequences of 12 protein-coding genes (PCGs) of acanthocephalan mitogenomes. Rotaria rotatoria and Philodina citrina are treated as the out-group. Bootstrap values ≥ 75 and Bayesian posterior probabilities values ≥ 0.95 are shown in the phylogenetic tree. Micracanthorhynchina hemirhamphi and Rhadinorhynchus laterospinosus are indicated using asterisk (*)