Introduction: Chromobacterium haemolyticum (C. haemolyticum) can cause invasive infections in humans. This study aims to reveal the genomic characteristics of C. haemolyticum and provide guidance for clinical diagnosis, treatment, prevention, and control.
Methods: Species identification was performed through isolation culture and matrix-assisted laser desorption ionization time-of-flight mass spectrometry. Antibiotic susceptibility testing determined resistance phenotypes. High-throughput sequencing and bioinformatics methods were used to predict antibiotic resistance genes and virulence genes and to analyze the evolutionary characteristics of global C. hemolyticus genomes.
Results: In this study, a C. haemolyticum strain was isolated from the bronchoalveolar lavage fluid of a patient in Guangxi Zhuang Autonomous Region, China. The isolate was sensitive to chloramphenicol, macrolides, and trimethoprim, while resistant to beta-lactams. Comparative genomics analysis revealed that most global strains carry carbapenemase-encoding genes. Phylogenetic analysis showed that the strain from this patient was closely related to a pond-derived C. haemolyticum isolate from Yangzhou, China.
Conclusions: This study uncovered the genetic characteristics of C. haemolyticum from various sources worldwide, including antibiotic resistance and virulence factors, providing an important reference for clinical treatment.
The genus Chromobacterium belongs to the family Neisseriaceae and comprises 19 species (1). Chromobacterium violaceum (C. violaceum) is a zoonotic pathogen found in tropical and subtropical regions that can cause severe sepsis with high mortality rates in humans (2). Since the first report of C. haemolyticum in 2008 (3), most invasive infection cases (e.g., pneumonia and bacteremia) have been associated with exposure to water bodies (4). However, genomic data on C. haemolyticum remains insufficient worldwide.
Here, we report the first case of pulmonary infection caused by Chromobacterium spp. in Guigang City, Guangxi Zhuang Autonomous Region, China. On the evening of November 4, 2023, an 18-year-old patient was admitted to the Qintang District People’s Hospital following a traffic accident in Guigang City. The patient subsequently developed pneumonia and a Chromobacterium spp. strain was isolated from bronchoalveolar lavage fluid. After combined treatment with cefoperazone sodium/sulbactam sodium, meropenem, and levofloxacin, the patient recovered.
Antibiotic susceptibility testing (AST) was conducted according to the Clinical and Laboratory Standards Institute (CLSI) guidelines for non-Enterobacteriaceae bacteria to determine the minimum inhibitory concentration (MIC) of the C. haemolyticum strain (5). The AST results are presented in Table 1. Overall, the strain demonstrated sensitivity to most antibiotics tested while exhibiting resistance to several beta-lactam and aminoglycoside antibiotics (Table 1 and
Table 1.
Results of antibiotic susceptibility testing.
The isolate was initially identified as C. violaceum using blood agar culture, biochemical experiments, and matrix-assisted laser desorption ionization time-of-flight mass spectrometry (MALDI-TOF MS) (model: VITEK MS, version: VITEK MS V3.0, BioMérieux, France). Due to similar cultural characteristics between the two species, C. haemolyticum is frequently misidentified as C. violaceum (6). Definitive identification as C. haemolyticum was subsequently achieved (
We downloaded 19 publicly available genomes of C. haemolyticum from the National Center for Biotechnology Information (NCBI) to characterize genomic features. After excluding two low-quality genomes (GCF_000285415.1 and GCF_003332145.1), the remaining 17 genomic datasets were used for subsequent analysis (
We detected 9 virulence factors (VFs), with 33.33% (3/9) belonging to the type 3 secretion system (
To explore the population evolution of C. haemolyticum, we analyzed 18 public C. haemolyticum genomes and 2 C. violaceum genomes (8). The phylogenetic tree revealed two distinct lineages corresponding to the two different species, spanning five countries and four diverse sources (human, water, environment, and Aedes aegypti) (Figure 1). Lineage one (L1) consisted of 2 C. violaceum strains from China, while lineage two (L2) comprised global C. haemolyticum strains. The isolate from the patient in this study showed a close genetic relationship with a pond-source C. haemolyticum strain from Yangzhou, China. Both isolates exhibited fewer virulence factors, with 6 VFs each.
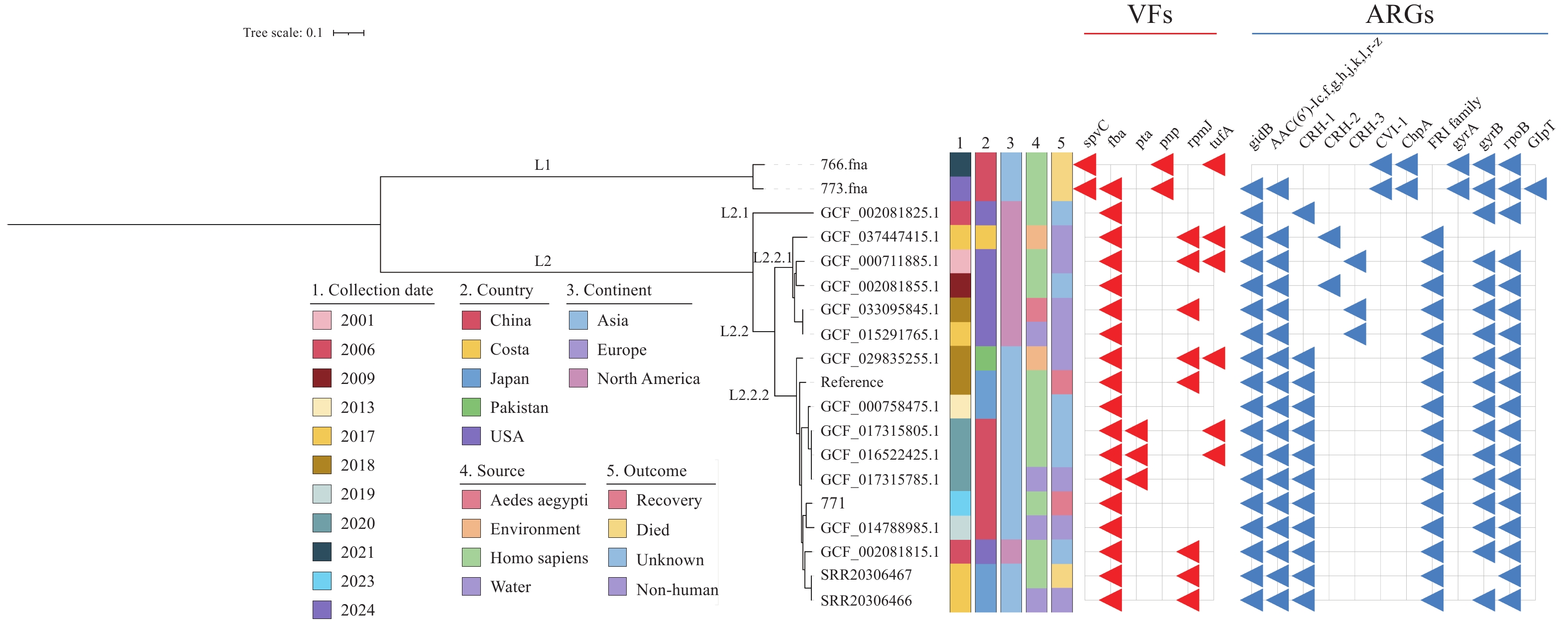
Phylogenetic analysis of 19 C. haemolyticum genomes.
Note: Phylogenetic tree-built details and analysis are described in the text. Collection date, country, source, continent, and outcome are labeled with different colors. The red and blue triangles represent the existence of VFs and ARGs, respectively.
Abbreviation: VFs=virulence factors; ARGs=antibiotic resistance genes.