Pena JC, de Kamino C, Rodrigues LHY, Mariano-Neto M, de Siqueira E. MF. Assessing the conservation status of species with limited available data and disjunct distribution. Biol Conserv. 2014;170:130–6. https://doi.org/10.1016/j.biocon.2013.12.015.
Meseguer AS, Lobo JM, Ree R, Beerling DJ, Sanmartín I. Integrating fossils, phylogenies, and niche models into biogeography to reveal ancient evolutionary history: the case of Hypericum (Hypericaceae). Syst Biol. 2015. https://doi.org/10.1093/sysbio/syu088
Google Scholar
Murphy SJ, Smith AB. What can community ecologists learn from species distribution models? Ecosphere. 2021. https://doi.org/10.1002/ecs2.3864.
Google Scholar
Jones LN, Leaché AD, Burbrink FT. Biogeographic barriers and historic climate shape the phylogeography and demography of the common gartersnake. J Biogeogr. 2023. https://doi.org/10.1111/jbi.14709.
Google Scholar
Jeon JY, Shin Y, Mularo AJ, Feng X, DeWoody JA. The integration of whole-genome resequencing and ecological niche modelling to conserve profiles of local adaptation. Divers Distrib. 2024;30:e13847.
Fick SE, Hijmans RJ. Worldclim 2: new 1-km spatial resolution climate surfaces for global land areas. Int J Climatol. 2017. https://doi.org/10.1002/joc.5086.
Google Scholar
Karger DN, Conrad O, Böhner J, Kawohl T, Kreft H, Soria-Auza RW, et al. Climatologies at high resolution for the earth’s land surface areas. Sci Data. 2017. https://doi.org/10.1038/sdata.2017.122.
Google Scholar
Title PO, Bemmels JB. Envirem: an expanded set of bioclimatic and topographic variables increases flexibility and improves performance of ecological niche modeling. Ecography. 2018. https://doi.org/10.1111/ecog.02880.
Google Scholar
Boria RA, Olson LE, Goodman SM, Anderson RP. Spatial filtering to reduce sampling bias can improve the performance of ecological niche models. Ecol Modell. 2014. https://doi.org/10.1016/j.ecolmodel.2013.12.012.
Google Scholar
Varela S, Anderson RP, García-Valdés R, Fernández-González F. Environmental filters reduce the effects of sampling bias and improve predictions of ecological niche models. Ecography. 2014. https://doi.org/10.1111/j.1600-0587.2013.00441.x.
Google Scholar
Warren DL, Wright AN, Seifert SN, Shaffer HB. Incorporating model complexity and spatial sampling bias into ecological niche models of climate change risks faced by 90 California vertebrate species of concern. Divers Distrib. 2014. https://doi.org/10.1111/ddi.12160.
Google Scholar
Anderson RP. Harnessing the world’s biodiversity data: promise and peril in ecological niche modeling of species distributions. Ann N Y Acad Sci. 2012. https://doi.org/10.1111/j.1749-6632.2011.06440.x.
Google Scholar
Botella C, Joly A, Monestiez P, Bonnet P, Munoz F. Bias in presence-only niche models related to sampling effort and species niches: lessons for background point selection. PLoS ONE. 2020;15:e0232078. https://doi.org/10.1371/journal.pone.0232078.
Moudrý V, Bazzichetto M, Remelgado R, Devillers R, Lenoir J, Mateo RG, et al. Optimising occurrence data in species distribution models: sample size, positional uncertainty, and sampling bias matter. Ecography. 2024. https://doi.org/10.1111/ecog.07294.
Google Scholar
Baker DJ, Hartley AJ, Butchart SHM, Willis SG. Choice of baseline climate data impacts projected species’ responses to climate change. Glob Chang Biol. 2016;22:2392–404. https://doi.org/10.1111/gcb.13273.
Bobrowski M, Udo S. Why input matters: selection of climate data sets for modelling the potential distribution of a treeline species in the Himalayan region. Ecol Modell. 2017;359:92–102. https://doi.org/10.1016/j.ecolmodel.2017.05.021.
Dubos N, Fieldsend TW, Roesch MA, Augros S, Besnard A, Choeur A et al. Choice of climate data influences predictions for current and future global invasion risks for two Phelsuma geckos. Biol Invasions. 2023;25:2929–48. https://doi.org/10.1007/s10530-023-03082-8.
Haesen S, Lenoir J, Gril E, De Frenne P, Lembrechts JJ, Kopecký M, et al. Microclimate reveals the true thermal niche of forest plant species. Ecol Lett. 2023. https://doi.org/10.1111/ele.14312.
Google Scholar
Kearney M, Porter W. Mechanistic niche modelling: combining physiological and spatial data to predict species’ ranges. Ecol Lett. 2009. https://doi.org/10.1111/j.1461-0248.2008.01277.x.
Google Scholar
Briscoe NJ, Morris SD, Mathewson PD, Buckley LB, Jusup M, Levy O, et al. Mechanistic forecasts of species responses to climate change: the promise of biophysical ecology. Glob Change Biol. 2023. https://doi.org/10.1111/gcb.16557.
Google Scholar
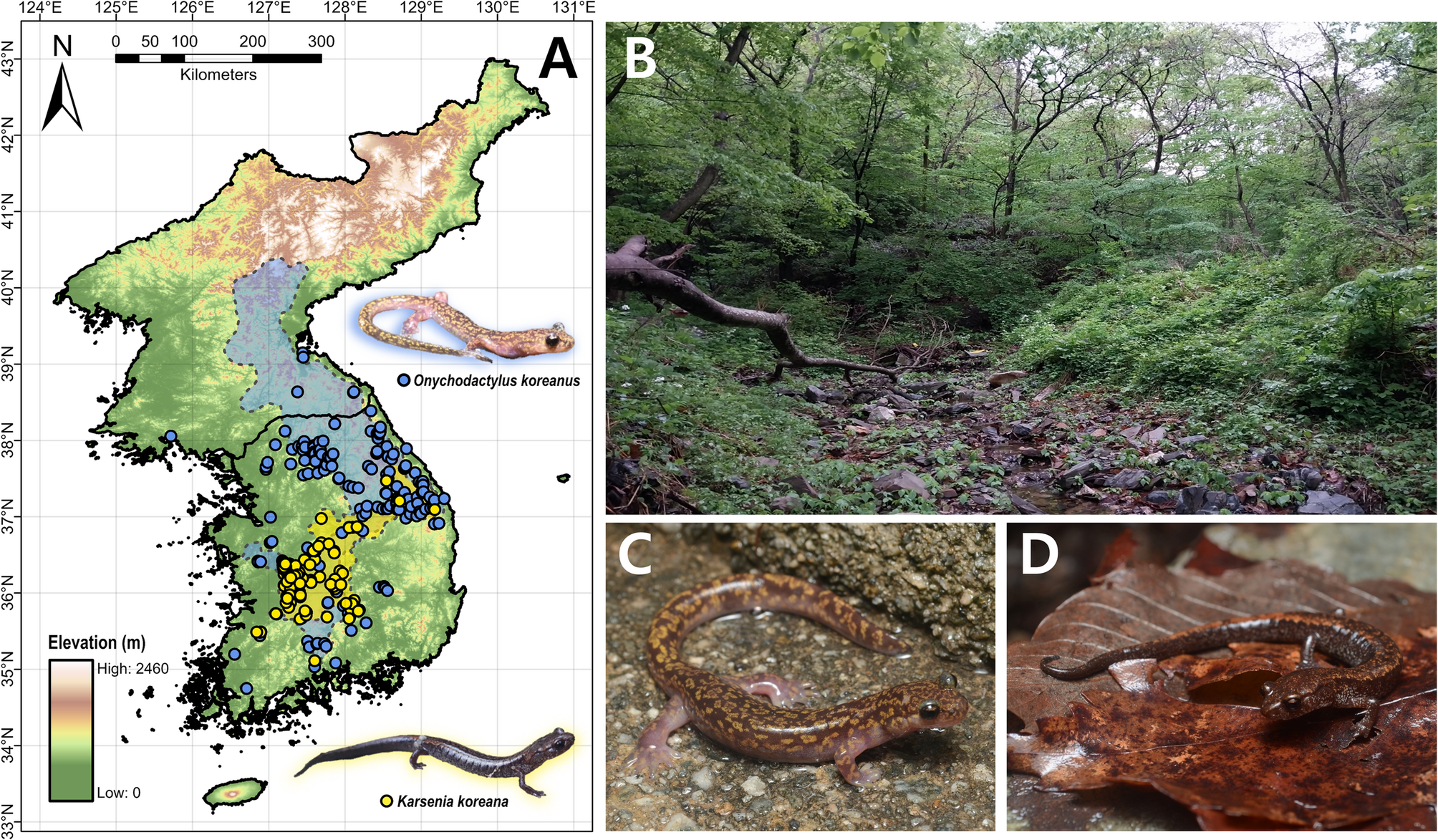
Lee JH, Ra NY, Eom J, Park D. Population dynamics of the long-tailed clawed salamander larva, Onychodactylus fischeri, and its age structure in Korea. J Ecol Field Biology. 2008;31:31–6.
Poyarkov NA, Che J, Min MS, Kuro-O M, Yan F, Li C et al. Review of the systematics, morphology and distribution of Asian clawed salamanders, genus Onychodactylus (Amphibia, caudata: Hynobiidae), with the description of four new species. Zootaxa. 2012:3465:1–106. https://doi.org/10.11646/zootaxa.3465.1.1.
Suk HY, Lee MY, Bae HG, Lee SJ, Poyarkov N, Lee H, et al. Phylogenetic structure and ancestry of Korean clawed salamander, Onychodactylus koreanus (Caudata: Hynobiidae). Mitochondr DNA A DNA Mapp Seq Anal. 2018. https://doi.org/10.1080/24701394.2017.1339187
Shin Y, Min MS, Borzée A. Driven to the edge: species distribution modeling of a clawed salamander (Hynobiidae: Onychodactylus koreanus) predicts range shifts and drastic decrease of suitable habitats in response to climate change. Ecol Evol. 2021;11:14669–88.
Google Scholar
Min MS, Yang SY, Bonett RM, Vieites DR, Brandon RA, Wake DB. Discovery of the first Asian plethodontid salamander. Nature. 2005. https://doi.org/10.1038/nature03474.
Google Scholar
Moon KY, Park D. Report of Karsenia koreana eggs oviposited within a semi-natural terrarium constructed at natural habitat. Korean J Herpetol. 2016;7:1–5.
Lee J-H, Park D. The encyclopedia of Korean amphibians. Seoul, South Korea: Nature and Ecology; 2016.
Borzée A, Litvinchuk SN, Ri K, Andersen D, Nam TY, Jon GH, et al. Update on distribution and conservation status of amphibians in the Democratic People’s Republic of Korea: conclusions based on field surveys, environmental modelling, molecular analyses and call properties. Animals. 2021. https://doi.org/10.3390/ani11072057.
Google Scholar
Vieites DR, Min MS, Wake DB. Rapid diversification and dispersal during periods of global warming by plethodontid salamanders. Proc Natl Acad Sci U S A. 2007. https://doi.org/10.1073/pnas.0705056104.
Google Scholar
Shen XX, Liang D, Chen MY, Mao RL, Wake DB, Zhang P. Enlarged multilocus data set provides surprisingly younger time of origin for the plethodontidae, the largest family of salamanders. Syst Biol. 2016;65:66–81. https://doi.org/10.1093/sysbio/syv061.
Jeon JY, Jung Jhwa, Suk HY, Lee H, Min MS. The Asian plethodontid salamander preserves historical genetic imprints of recent Northern expansion. Sci Rep. 2021. https://doi.org/10.1038/s41598-021-88238-z.
Google Scholar
Borzée A, Andersen D, Groffen J, Kim HT, Bae Y, Jang Y. Climate change-based models predict range shifts in the distribution of the only Asian plethodontid salamander: Karsenia koreana. Sci Rep. 2019. https://doi.org/10.1038/s41598-019-48310-1.
Google Scholar
Hewitt G. The genetic legacy of the quaternary ice ages. Nature. 2000. https://doi.org/10.1038/35016000.
Google Scholar
Kim HW, Yoon S, Kim M, Shin M, Yoon H, Kim K. EcoBank: A flexible database platform for sharing ecological data. Biodivers Data J. 2021;9:e61866. https://doi.org/10.3897/BDJ.9.e61866.
Shipley BR, Bach R, Do Y, Strathearn H, McGuire JL, Dilkina B. megaSDM: integrating dispersal and time-step analyses into species distribution models. Ecography. 2022;2022:e05450. https://doi.org/10.1111/ecog.05450.
Vignali S, Barras AG, Arlettaz R, Braunisch V. SDMtune: an R package to tune and evaluate species distribution models. Ecol Evol. 2020;10:11488–506. https://doi.org/10.1002/ece3.6786.
R Core Team. R: A language and environment for statistical computing, Version 4.2.2. 2022. https://www.Rproject.org//. Accessed 19 Sep 2024.
Venables WN, Ripley BD. Modern Applied Statistics with S. 4th Edition. New York: Springer; 2002.
Tuanmu MN, Jetz W. A global 1-km consensus land-cover product for biodiversity and ecosystem modelling. Glob Ecol Biogeogr. 2014. https://doi.org/10.1111/geb.12182.
Google Scholar
Hijmans RJ. raster: Geographic Data Analysis and Modeling. 2023.
Peng Y, Li Y, Cao G, Li H, Shin Y, Piao Z, et al. Estimation of habitat suitability and landscape connectivity for Liaoning and Jilin clawed salamanders (Hynobiidae: Onychodactylus) in the transboundary region between the People’s Republic of China and the Democratic People’s Republic of Korea. Glob Ecol Conserv. 2023. https://doi.org/10.1016/j.gecco.2023.e02694.
Google Scholar
Borzée A. Continental Northeast Asian Amphibians Origins, Behavioural Ecology, and Conservation. 1st edition. Amsterdam: Elsevier; 2024.
Osorio-Olvera L, Lira-Noriega A, Soberón J, Peterson AT, Falconi M, Contreras-Díaz RG, et al. Ntbox: an R package with graphical user interface for modelling and evaluating multidimensional ecological niches. Methods Ecol Evol. 2020. https://doi.org/10.1111/2041-210X.13452.
Google Scholar
Phillips SJ, Anderson RP, Dudík M, Schapire RE, Blair ME. Opening the black box: an open-source release of maxent. Ecography. 2017. https://doi.org/10.1111/ecog.03049.
Google Scholar
Kass JM, Muscarella R, Galante PJ, Bohl CL, Pinilla-Buitrago GE, Boria RA, et al. ENMeval 2.0: redesigned for customizable and reproducible modeling of species’ niches and distributions. Methods Ecol Evol. 2021. https://doi.org/10.1111/2041-210X.13628.
Google Scholar
Roberts DR, Bahn V, Ciuti S, Boyce MS, Elith J, Guillera-Arroita G, et al. Cross-validation strategies for data with temporal, spatial, hierarchical, or phylogenetic structure. Ecography. 2017. https://doi.org/10.1111/ecog.02881.
Google Scholar
Valavi R, Elith J, Lahoz-Monfort JJ, Guillera-Arroita G, blockCV. An r package for generating spatially or environmentally separated folds for k-fold cross-validation of species distribution models. Methods Ecol Evol. 2019;10:225–32. https://doi.org/10.1111/2041-210X.13107.
Muscarella R, Galante PJ, Soley-Guardia M, Boria RA, Kass JM, Uriarte M et al. ENMeval: an R package for conducting spatially independent evaluations and estimating optimal model complexity for maxent ecological niche models. Methods Ecol Evol. 2014;5:1198–205. https://doi.org/10.1111/2041-210X.12261.
Melton AE, Clinton MH, Wasoff DN, Lu L, Hu H, Chen Z, et al. Climatic niche comparisons of Eastern North American and Eastern Asian disjunct plant genera. Glob Ecol Biogeogr. 2022. https://doi.org/10.1111/geb.13504.
Google Scholar
Warren DL, Seifert SN. Ecological niche modeling in maxent: the importance of model complexity and the performance of model selection criteria. Ecol Appl. 2011. https://doi.org/10.1890/10-1171.1.
Google Scholar
Radosavljevic A, Anderson RP. Making better maxent models of species distributions: complexity, overfitting and evaluation. J Biogeogr. 2014. https://doi.org/10.1111/jbi.12227.
Google Scholar
Low BW, Zeng Y, Tan HH, Yeo DCJ. Predictor complexity and feature selection affect maxent model transferability: evidence from global freshwater invasive species. Divers Distrib. 2021. https://doi.org/10.1111/ddi.13211.
Google Scholar
Lobo JM, Jiménez-valverde A, Real R. AUC: a misleading measure of the performance of predictive distribution models. Glob Ecol Biogeogr. 2008. https://doi.org/10.1111/j.1466-8238.2007.00358.x.
Google Scholar
Jiménez-Valverde A. Insights into the area under the receiver operating characteristic curve (AUC) as a discrimination measure in species distribution modelling. Glob Ecol Biogeogr. 2012. https://doi.org/10.1111/j.1466-8238.2011.00683.x.
Google Scholar
Hirzel AH, Le Lay G, Helfer V, Randin C, Guisan A. Evaluating the ability of habitat suitability models to predict species presences. Ecol Modell. 2006. https://doi.org/10.1016/j.ecolmodel.2006.05.017.
Google Scholar
Bohl CL, Kass JM, Anderson RP. A new null model approach to quantify performance and significance for ecological niche models of species distributions. J Biogeogr. 2019. https://doi.org/10.1111/jbi.13573.
Google Scholar
Zurell D, Franklin J, König C, Bouchet PJ, Dormann CF, Elith J et al. A standard protocol for reporting species distribution models. Ecography. 2020;43:1261–77. https://doi.org/10.1111/ecog.04960.
Fitzpatrick MC, Lachmuth S, Haydt NT. The ODMAP protocol: a new tool for standardized reporting that could revolutionize species distribution modeling. Ecography. 2021. https://doi.org/10.1111/ecog.05700.
Google Scholar
Otto-Bliesner BL, Marshall SJ, Overpeck JT, Miller GH, Hu A. Simulating arctic climate warmth and icefield retreat in the last interglaciation. Science. 2006;311:1751–53. https://doi.org/10.1126/science.1120808.
Fordham DA, Saltré F, Haythorne S, Wigley TML, Otto-Bliesner BL, Chan KC, et al. Paleoview: a tool for generating continuous climate projections spanning the last 21 000 years at regional and global scales. Ecography. 2017. https://doi.org/10.1111/ecog.03031.
Google Scholar
Brown JL, Hill DJ, Dolan AM, Carnaval AC, Haywood AM. Paleoclim, high Spatial resolution paleoclimate surfaces for global land areas. Sci Data. 2018;5:180254.https://doi.org/10.1038/sdata.2018.254.
Elith J, Kearney M, Phillips S. The Art of modelling range-shifting species. Methods Ecol Evol. 2010;1:330–42. https://doi.org/10.1111/j.2041-210X.2010.00036.x.
Dubos N, Augros S, Deso G, Probst JM, Notter JC, Roesch MA. Here be dragons: important Spatial uncertainty driven by climate data in forecasted distribution of an endangered insular reptile. Anim Conserv. 2022;25:704–17. https://doi.org/10.1111/acv.12775.
Schoener TW. The Anolis lizards of bimini: resource partitioning in a complex fauna. Ecology. 1968. https://doi.org/10.2307/1935534.
Google Scholar
Warren DL, Glor RE, Turelli M. Environmental niche equivalency versus conservatism: quantitative approaches to niche evolution. Evolution. 2008. https://doi.org/10.1111/j.1558-5646.2008.00482.x.
Google Scholar
Warren DL, Matzke NJ, Cardillo M, Baumgartner JB, Beaumont LJ, Turelli M, et al. ENMTools 1.0: an R package for comparative ecological biogeography. Ecography. 2021. https://doi.org/10.1111/ecog.05485.
Google Scholar
Broennimann O, Fitzpatrick MC, Pearman PB, Petitpierre B, Pellissier L, Yoccoz NG, et al. Measuring ecological niche overlap from occurrence and spatial environmental data. Glob Ecol Biogeogr. 2012. https://doi.org/10.1111/j.1466-8238.2011.00698.x.
Google Scholar
Brown JL, Carnaval AC. A Tale of two niches: methods, concepts, and evolution. Front Biogeogr. 2019;11:e44158. https://doi.org/10.21425/F5FBG44158.
Brown JL. humboldt R package documentation. 2019. https://jasonleebrown.github.io/humboldt/. Accessed 8 Mar 2025.
Roubicek AJ, VanDerWal J, Beaumont LJ, Pitman AJ, Wilson P, Hughes L. Does the choice of climate baseline matter in ecological niche modelling? Ecol Modell. 2010. https://doi.org/10.1016/j.ecolmodel.2010.06.021.
Google Scholar
Nishikawa K. The first specimen of Karsenia Koreana (Caudata: Plethodontidae) collected 34 years before its description. Curr Herpetol. 2009;28:27–8. https://doi.org/10.3105/018.028.0104.
Shin Y, Jang Y, Kim T, Borzée A. A specimen of Karsenia koreana (caudata: plethodontidae) misidentified as Hynobius leechii 27 years before the species’ description and additional historical record. Curr Herpetol. 2020;39:75–9. https://doi.org/10.5358/hsj.39.75.
Guillon M, Martínez-Freiría F, Lucchini N, Ursenbacher S, Surget-Groba Y, Kageyama M, et al. Inferring current and last glacial maximum distributions are improved by physiology-relevant climatic variables in cold-adapted ectotherms. J Biogeogr. 2024. https://doi.org/10.1111/jbi.14828.
Google Scholar
Kim S-J, Park Y-M, Lee B-Y, Choi T-J, Yoon Y-J, Suk B-C. Study of East Asia climate change for the last glacial maximum using numerical model. Korean J Quaternary Res. 2006;20:51–66.
Kim SJ, Kim JW, Kim BM. Last glacial maximum climate over Korean Peninsula in PMIP3 simulations. Quat Int. 2015. https://doi.org/10.1016/j.quaint.2015.02.062.
Google Scholar
Escoriza D, Hassine JB. Niche partitioning at local and regional scale in the North African Salamandridae. J Herpetol. 2015;49:276–83. https://doi.org/10.1670/13-151.
Barbet-Massin M, Rome Q, Villemant C, Courchamp F. Can species distribution models really predict the expansion of invasive species? PLoS One. 2018. https://doi.org/10.1371/journal.pone.0193085.
Google Scholar
Chiarenza AA, Mannion PD, Lunt DJ, Farnsworth A, Jones LA, Kelland SJ et al. Ecological niche modelling does not support climatically-driven dinosaur diversity decline before the cretaceous/paleogene mass extinction. Nat Commun. 2019;10:1091. https://doi.org/10.1038/s41467-019-08997-2.
Feng X, Park DS, Walker C, Peterson AT, Merow C, Papeş M. A checklist for maximizing reproducibility of ecological niche models. Nat Ecol Evol. 2019. https://doi.org/10.1038/s41559-019-0972-5.
Google Scholar
Riddell EA, Odom JP, Damm JD, Sears MW. Plasticity reveals hidden resistance to extinction under climate change in the global hotspot of salamander diversity. Sci Adv. 2018. https://doi.org/10.1126/sciadv.aar5471.
Google Scholar
Lyons MP, Kozak KH. Vanishing islands in the sky? A comparison of correlation- and mechanism-based forecasts of range dynamics for montane salamanders under climate change. Ecography. 2020. https://doi.org/10.1111/ecog.04282.
Google Scholar
Riddell E, Sears MW. Terrestrial salamanders maintain habitat suitability under climate change despite trade-offs between water loss and gas exchange. Physiol Biochem Zool. 2020. https://doi.org/10.1086/709558.
Google Scholar
Velazco SJE, Rose MB, de Andrade AFA, Minoli I, Franklin J. Flexsdm: an r package for supporting a comprehensive and flexible species distribution modelling workflow. Methods Ecol Evol. 2022;13:1661–9. https://doi.org/10.1038/s41467-019-08997-2.
Thuiller W, Lafourcade B, Engler R, Araújo MB. Biomod – a platform for ensemble forecasting of species distributions. Ecography. 2009. https://doi.org/10.1111/j.1600-0587.2008.05742.x.
Google Scholar
Van Aelst S, Rousseeuw P. Minimum volume ellipsoid. Wiley Interdisciplinary Reviews: Comput Stat. 2009;1:71–82. https://doi.org/10.1002/wics.19.
Ficetola GF, Lunghi E, Canedoli C, Padoa-Schioppa E, Pennati R, Manenti R. Differences between microhabitat and broad-scale patterns of niche evolution in terrestrial salamanders. Sci Rep. 2018. https://doi.org/10.1038/s41598-018-28796-x.
Google Scholar
Jung J-H. Habitat characteristics of three salamanders (Caudata: Amphibia) in forests and genetic diversity of Karsenia koreana. PhD Dissertation. Seoul National University; 2020.
Oboudi R, Malekian M, Khosravi R, Fadakar D, Adibi MA. Genetic structure and ecological niche segregation of Indian Gray mongoose (Urva edwardsii) in Iran. Ecol Evol. 2021. https://doi.org/10.1002/ece3.8168.
Google Scholar
Niwa K, Tran D, Van, Nishikawa K. Differentiated historical demography and ecological niche forming present distribution and genetic structure in coexisting two salamanders (Amphibia, urodela, Hynobiidae) in a small island, Japan. PeerJ. 2022;10:e13202.
Google Scholar
Tran D, Van, Tominaga A, Pham LT, Nishikawa K. Ecological niche modeling shed light on new insights of the speciation processes and historical distribution of Japanese fire-bellied Newt Cynops pyrrhogaster (Amphibia: Urodela). Ecol Inf. 2024;79:102443. https://doi.org/10.1016/j.ecoinf.2023.102443.
Pyron RA, Pirro S, Hains T, Colston TJ, Myers EA, O’Connell KA et al. The draft genome sequences of 50 salamander species (Caudata, Amphibia). Biodiversity Genomes. 2024. https://doi.org/10.56179/001c.116891.
Pyron RA, O’Connell KA, Lemmon EM, Lemmon AR, Beamer DA. Phylogenomic data reveal reticulation and incongruence among mitochondrial candidate species in Dusky salamanders (Desmognathus). Mol Phylogenet Evol. 2020;146:106751. https://doi.org/10.1016/j.ympev.2020.106751.
Burgon JD, Vences M, Steinfartz S, Bogaerts S, Bonato L, Donaire-Barroso D et al. Phylogenomic inference of species and subspecies diversity in the Palearctic salamander genus Salamandra. Mol Phylogenet Evol. 2021;157:107063. https://doi.org/10.1016/j.ympev.2020.107063.
Pyron RA, O’connell KA, Duncan SC, Burbrink FT, Beamer DA. Speciation hypotheses from phylogeographic delimitation yield an integrative taxonomy for seal salamanders (Desmognathus monticola). Syst Biol. 2023. https://doi.org/10.1093/sysbio/syac065.
Google Scholar
Talavera A, Palmada-Flores M, Burriel-Carranza B, Valbuena-Ureña E, Mochales-Riaño G, Adams DC, et al. Genomic insights into the Montseny brook newt (Calotriton arnoldi), a critically endangered glacial relict. iScience. 2024. https://doi.org/10.1016/j.isci.2023.108665.
Google Scholar
Borzée A, Shin Y, Poyarkov NA, Jeon JY, Baek HJ, Lee CH, et al. Dwindling in the mountains: description of a critically endangered and microendemic Onychodactylus species (Amphibia, Hynobiidae) from the Korean Peninsula. Zool Res. 2022;43:750–5. https://doi.org/10.24272/j.issn.2095-8137.2022.048.
Rhoden CM, Peterman WE, Taylor CA. Maxent-directed field surveys identify new populations of narrowly endemic habitat specialists. PeerJ. 2017;5:e3632. https://doi.org/10.7717/peerj.3632.
Sakai Y, Kusakabe A, Tsuchida K, Tsuzuku Y, Okada S, Kitamura T, et al. Discovery of an unrecorded population of Yamato salamander (Hynobius vandenburghi) by GIS and edna analysis. Environ DNA. 2019. https://doi.org/10.1002/edn3.31.
Google Scholar