Mimicking the way experienced human observers track growth over time, 3D-NOD integrates novel labeling, registration, and data augmentation strategies to boost sensitivity and accuracy. Tested across multiple crop species, the system achieved an impressive mean F1-score of 88.13% and IoU of 80.68%, offering a powerful tool for real-time, organ-level plant phenotyping.
Accurate plant growth monitoring underpins modern agriculture, enabling yield prediction, stress detection, and precise phenotyping. Traditional methods—measuring plant height, canopy volume, or detecting diseases—often rely on 2D images, which cannot fully capture depth or resolve self-occluded structures. Spatiotemporal phenotyping, which tracks individual organs over time, offers richer insights but faces challenges in detecting small, newly emerged organs and handling complex plant architectures. 3D sensing technology addresses these limitations by capturing depth information, but current 3D approaches are either computationally heavy or require organs to be large enough for tracking. Based on these challenges, the researchers developed a method to improve early-stage detection of plant growth events from time-series 3D data.
A study (DOI: 10.1016/j.plaphe.2025.100002) published in Plant Phenomics on 22 February 2025 by Dawei Li’s team, Donghua University, presents a highly sensitive 3D deep learning framework for detecting new plant organs, enabling more accurate and real-time growth monitoring to advance precision agriculture and phenotyping.
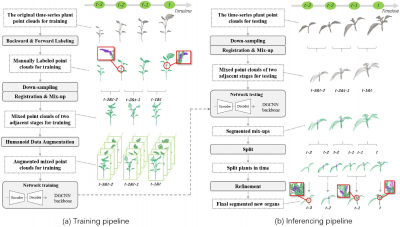
The researchers evaluated the 3D-NOD framework using a high-precision 3D plant dataset containing tobacco, tomato, and sorghum. They constructed a spatiotemporal dataset of 37 growth sequences, comprising 468 point clouds, each with over ten growth stages. Since most sequences captured seedlings, the primary growth events were budding. Using the Semantic Segmentation Editor under Ubuntu, they annotated all points under the Backward & Forward Labeling (BFL) strategy into two semantic classes—“old organ” and “new organ.” The training set included 25 sequences and the test set 12 sequences. To enhance learning, each mixed point cloud underwent Humanoid Data Augmentation (HDA) to generate ten variants for training the DGCNN backbone. Performance was assessed with Precision, Recall, F1-score, and Intersection over Union (IoU). Compared to PointNet, PointNet++, DGCNN, and PAConv, 3D-NOD achieved superior sensitivity in new organ detection, with F1 and IoU for new organs reaching 76.65% and 62.14%, respectively, despite many buds being too small for human identification. Qualitative results confirmed accurate detection of tiny buds across all three species with low false alarms. Ablation studies demonstrated that removing any key component—BFL, Registration & Mix-up (RMU), or parts of HDA—caused noticeable performance declines, underscoring their combined importance. Interestingly, detection was best in sorghum, likely due to its faster bud growth. To meet real-time phenotyping needs, the team adapted the pipeline for single point cloud testing by creating pseudo-temporal inputs, enabling inference without multiple growth stages. Comparative analyses on tomato, tobacco, and sorghum sequences showed only minor accuracy reductions compared to standard spatiotemporal testing, highlighting the framework’s versatility for both multi-stage and single-stage growth monitoring scenarios.
3D-NOD offers a robust, scalable solution for precision agriculture and breeding programs. By accurately detecting organ-level growth events early, it enables researchers and growers to monitor crop development more closely, improve trait measurements, and optimize resource use. This capability is vital for high-throughput phenotyping, where rapid and non-invasive measurement of plant traits drives selection efficiency. The framework’s adaptability to different species suggests broad potential in monitoring diverse crops under field or greenhouse conditions. Beyond agriculture, the method could inform botanical research, forestry, and ecological studies where fine-scale structural changes in plants are critical indicators of health and development.
###
References
DOI
10.1016/j.plaphe.2025.100002
Original Source URL
https://doi.org/10.1016/j.plaphe.2025.100002
Funding information
This work was supported by the self-collected funds from Dawei Li.
About Plant Phenomics
Science Partner Journal Plant Phenomics is an online-only Open Access journal published in affiliation with the State Key Laboratory of Crop Genetics & Germplasm Enhancement, Nanjing Agricultural University (NAU) and distributed by the American Association for the Advancement of Science (AAAS). Like all partners participating in the Science Partner Journal program, Plant Phenomics is editorially independent from the Science family of journals. Editorial decisions and scientific activities pursued by the journal’s Editorial Board are made independently, based on scientific merit and adhering to the highest standards for accurate and ethical promotion of science. These decisions and activities are in no way influenced by the financial support of NAU, NAU administration, or any other institutions and sponsors. The Editorial Board is solely responsible for all content published in the journal. To learn more about the Science Partner Journal program, visit the SPJ program homepage.