Torre LA, Siegel RL, Ward EM, Jemal A. Global cancer incidence and mortality rates and Trends—An update. Cancer Epidemiol Biomarkers Prev. 2016;25(1):16–27.
Google Scholar
Siegel RL, Miller KD, Wagle NS, Jemal A. Cancer statistics, 2023. Cancer J Clin. 2023;73(1):17–48.
Heer E, Harper A, Escandor N, Sung H, McCormack V, Fidler-Benaoudia MM. Global burden and trends in premenopausal and postmenopausal breast cancer: a population-based study. Lancet Global Health. 2020;8(8):e1027–37.
Google Scholar
Torre LA, Islami F, Siegel RL, Ward EM, Jemal A. Global cancer in women: burden and trends. Cancer Epidemiol Biomarkers Prev. 2017;26(4):444–57.
Google Scholar
Kehm RD, Yang W, Tehranifar P, Terry MB. 40 years of change in Age- and Stage-Specific cancer incidence rates in US women and men. JNCI Cancer Spectr 2019;3(3).
Emilsson V, Ilkov M, Lamb JR, et al. Co-regulatory networks of human serum proteins link genetics to disease. Science. 2018;361(6404):769–73.
Google Scholar
Schwenk JM, Omenn GS, Sun Z, et al. The human plasma proteome draft of 2017: Building on the human plasma PeptideAtlas from mass spectrometry and complementary assays. J Proteome Res. 2017;16(12):4299–310.
Google Scholar
International Human Genome Sequencing C. Finishing the euchromatic sequence of the human genome. Nature. 2004;431(7011):931–45.
Lin H, Lee E, Hestir K, et al. Discovery of a cytokine and its receptor by functional screening of the extracellular proteome. Science. 2008;320(5877):807–11.
Google Scholar
Williams SA, Kivimaki M, Langenberg C, et al. Plasma protein patterns as comprehensive indicators of health. Nat Med. 2019;25(12):1851–7.
Google Scholar
Key TJ, Appleby PN, Reeves GK, Roddam AW. Insulin-like growth factor 1 (IGF1), IGF binding protein 3 (IGFBP3), and breast cancer risk: pooled individual data analysis of 17 prospective studies. Lancet Oncol. 2010;11(6):530–42.
Google Scholar
Mälarstig A, Grassmann F, Dahl L, et al. Evaluation of Circulating plasma proteins in breast cancer using Mendelian randomisation. Nat Commun. 2023;14(1):7680.
Google Scholar
Shu X, Bao J, Wu L, et al. Evaluation of associations between genetically predicted Circulating protein biomarkers and breast cancer risk. Int J Cancer. 2020;146(8):2130–8.
Google Scholar
Song J, Yang H. Identifying new biomarkers and potential therapeutic targets for breast cancer through the integration of human plasma proteomics: a Mendelian randomization study and colocalization analysis. Front Endocrinol (Lausanne). 2024;15:1449668.
Google Scholar
Papier K, Atkins JR, Tong TYN, et al. Identifying proteomic risk factors for cancer using prospective and exome analyses of 1463 Circulating proteins and risk of 19 cancers in the UK biobank. Nat Commun. 2024;15(1):4010.
Google Scholar
Grassmann F, Mälarstig A, Dahl L, et al. The impact of Circulating protein levels identified by affinity proteomics on short-term, overall breast cancer risk. Br J Cancer. 2024;130(4):620–7.
Google Scholar
Terry MB, Phillips KA, Daly MB, et al. Cohort profile: the breast cancer prospective family study cohort (ProF-SC). Int J Epidemiol. 2016;45(3):683–92.
Google Scholar
Pharoah P, Day N, Duffy S, Easton D, Ponder B. Family history and the risk of breast cancer: a systematic review and meta-analysis. Int J Cancer. 1997;71:800–9.
Google Scholar
Braithwaite D, Miglioretti DL, Zhu W, et al. Family history and breast cancer risk among older women in the breast cancer surveillance consortium cohort. JAMA Intern Med. 2018;178(4):494–501.
Google Scholar
Rinschen MM, Ivanisevic J, Giera M, Siuzdak G. Identification of bioactive metabolites using activity metabolomics. Nat Rev Mol Cell Biol. 2019;20(6):353–67.
Google Scholar
Wu HC, Lai Y, Liao Y, et al. Plasma metabolomics profiles and breast cancer risk. Breast Cancer Res. 2024;26(1):141.
Google Scholar
John E, Hopper J, Beck J, et al. The breast cancer family registry: an infrastructure for cooperative multinational, interdisciplinary and translational studies of the genetic epidemiology of breast cancer. Breast Cancer Res. 2004;6(4):R375–89.
Google Scholar
Uppal K, Ma C, Go YM, Jones DP, Wren J. xMWAS: a data-driven integration and differential network analysis tool. Bioinformatics. 2018;34(4):701–2.
Google Scholar
Terry MB, Phillips K-A, Daly MB et al. Cohort profile: the breast cancer prospective family study cohort (ProF-SC). Int J Epidemiol 2015:1–10.
Lundberg M, Eriksson A, Tran B, Assarsson E, Fredriksson S. Homogeneous antibody-based proximity extension assays provide sensitive and specific detection of low-abundant proteins in human blood. Nucleic Acids Res. 2011;39(15):e102.
Google Scholar
Assarsson E, Lundberg M, Holmquist G, et al. Homogenous 96-Plex PEA immunoassay exhibiting high sensitivity, specificity, and excellent scalability. PLoS ONE. 2014;9(4):e95192.
Google Scholar
Lee A, Mavaddat N, Wilcox AN, et al. BOADICEA: a comprehensive breast cancer risk prediction modelincorporating genetic and nongenetic risk factors. Genet Sci. 2019;21(8):1708–18.
Antoniou AC, Cunningham AP, Peto J, Evans DG, Lalloo F, Narod SA. The BOADICEA model of genetic susceptibility to breast and ovarian cancers: updates and extensions. Br J Cancer 2008;98.
Terry MB, Liao Y, Whittemore AS, et al. 10-year performance of four models of breast cancer risk: a validation study. Lancet Oncol. 2019;20(4):504–17.
Google Scholar
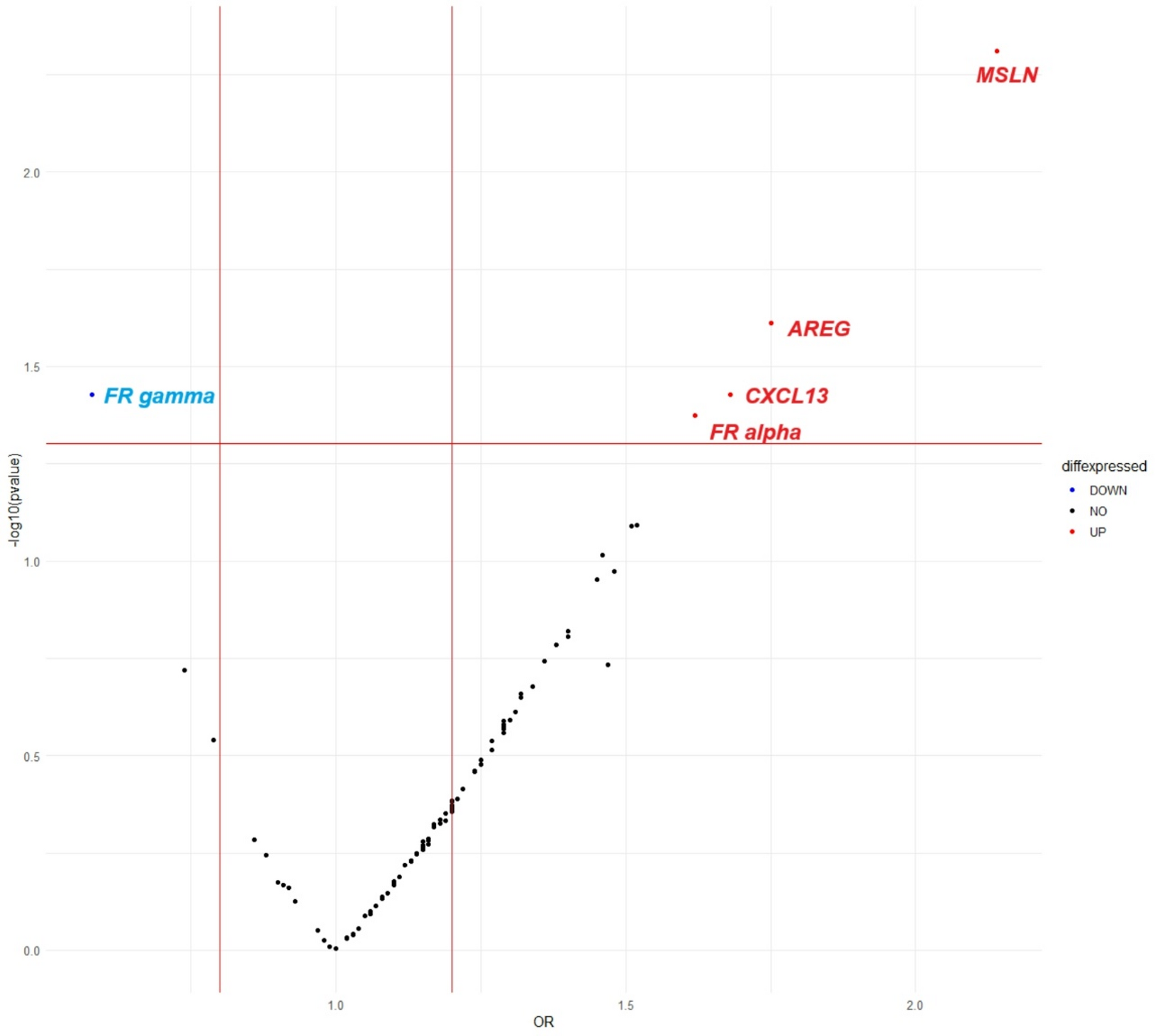
Chang K, Pastan I. Molecular cloning of mesothelin, a differentiation antigen present on mesothelium, mesotheliomas, and ovarian cancers. Proc Natl Acad Sci U S A. 1996;93(1):136–40.
Google Scholar
Chang K, Pastan I. Molecular cloning and expression of a cDNA encoding a protein detected by the K1 antibody from an ovarian carcinoma (OVCAR-3) cell line. Int J Cancer. 1994;57(1):90–7.
Google Scholar
Frierson HF, Moskaluk CA, Powell SM, et al. Large-scale molecular and tissue microarray analysis of mesothelin expression in common human carcinomas. Hum Pathol. 2003;34(6):605–9.
Google Scholar
Parinyanitikul N, Blumenschein GR, Wu Y, et al. Mesothelin expression and survival outcomes in triple receptor negative breast cancer. Clin Breast Cancer. 2013;13(5):378–84.
Google Scholar
Scholler N, Fu N, Yang Y, et al. Soluble member(s) of the mesothelin/megakaryocyte potentiating factor family are detectable in Sera from patients with ovarian carcinoma. Proc Natl Acad Sci. 1999;96(20):11531–6.
Google Scholar
Hassan R, Remaley AT, Sampson ML, et al. Detection and quantitation of serum mesothelin, a tumor marker for patients with mesothelioma and ovarian cancer. Clin Cancer Res. 2006;12(2):447–53.
Google Scholar
Saha S, Mukherjee C, Basak D, et al. High expression of mesothelin in plasma and tissue is associated with poor prognosis and promotes invasion and metastasis in gastric cancer. Adv Cancer Biology – Metastasis. 2023;7:100098.
Busser B, Sancey L, Brambilla E, Coll J-L, Hurbin A. The multiple roles of Amphiregulin in human cancer. Biochim Et Biophys Acta (BBA) – Reviews Cancer. 2011;1816(2):119–31.
Ciarloni L, Mallepell S, Brisken C. Amphiregulin is an essential mediator of Estrogen receptor alpha function in mammary gland development. Proc Natl Acad Sci U S A. 2007;104(13):5455–60.
Google Scholar
So WK, Fan Q, Lau MT, Qiu X, Cheng JC, Leung PC. Amphiregulin induces human ovarian cancer cell invasion by down-regulating E-cadherin expression. FEBS Lett. 2014;588(21):3998–4007.
Google Scholar
Willmarth NE, Ethier SP. Autocrine and juxtacrine effects of Amphiregulin on the proliferative, invasive, and migratory properties of normal and neoplastic human mammary epithelial cells. J Biol Chem. 2006;281(49):37728–37.
Google Scholar
Lofgren KA, Reker NC, Sreekumar S, Kenny PA. Pan-cancer distribution of cleaved cell-surface amphiregulin, the target of the GMF-1A3 antibody drug conjugate. Antib Ther. 2022;5(3):226–31.
Google Scholar
Kjær IM, Olsen DA, Brandslund I, et al. Dysregulated EGFR pathway in serum in early-stage breast cancer patients: A case control study. Sci Rep. 2020;10(1):6714.
Google Scholar
Panse J, Friedrichs K, Marx A, et al. Chemokine CXCL13 is overexpressed in the tumour tissue and in the peripheral blood of breast cancer patients. Br J Cancer. 2008;99(6):930–8.
Google Scholar
Chen L, Huang Z, Yao G, et al. The expression of CXCL13 and its relation to unfavorable clinical characteristics in young breast cancer. J Transl Med. 2015;13:168.
Google Scholar
Gunn MD, Ngo VN, Ansel KM, Ekland EH, Cyster JG, Williams LT. A B-cell-homing chemokine made in lymphoid follicles activates burkitt’s lymphoma receptor-1. Nature. 1998;391(6669):799–803.
Google Scholar
Scaranti M, Cojocaru E, Banerjee S, Banerji U. Exploiting the folate receptor α in oncology. Nat Reviews Clin Oncol. 2020;17(6):349–59.
Cheung A, Opzoomer J, Ilieva KM, et al. Anti-Folate receptor Alpha–Directed antibody therapies restrict the growth of Triple-negative breast cancer. Clin Cancer Res. 2018;24(20):5098–111.
Google Scholar
Cheung A, Bax HJ, Josephs DH et al. Targeting folate receptor alpha for cancer treatment. Oncotarget 2016;7(32).
Kelley KMM, Rowan BG, Ratnam M. Modulation of the folate receptor α gene by the Estrogen receptor: mechanism and implications in tumor Targeting12. Cancer Res. 2003;63(11):2820–8.
Google Scholar
Necela BM, Crozier JA, Andorfer CA, et al. Folate Receptor-α (FOLR1) expression and function in triple negative tumors. PLoS ONE. 2015;10(3):e0122209.
Google Scholar
Bax HJ, Chauhan J, Stavraka C, et al. Folate receptor alpha in ovarian cancer tissue and patient serum is associated with disease burden and treatment outcomes. Br J Cancer. 2023;128(2):342–53.
Google Scholar
Shen F, Wu M, Ross JF, Miller D, Ratnam M. Folate receptor type gamma is primarily a secretory protein due to lack of an efficient signal for glycosylphosphatidylinositol modification: protein characterization and cell type specificity. Biochemistry. 1995;34(16):5660–5.
Google Scholar
Viswanathan S, Parida S, Lingipilli BT, Krishnan R, Podipireddy DR, Muniraj N. Role of gut microbiota in breast cancer and drug resistance. Pathogens 2023;12(3).
Malinowska AM, Schmidt M, Kok DE, Chmurzynska A. Ex vivo folate production by fecal bacteria does not predict human blood folate status: associations between dietary patterns, gut microbiota, and folate metabolism. Food Res Int. 2022;156:111290.
Google Scholar
Lephart ED, Naftolin F. Estrogen action and gut Microbiome metabolism in dermal health. Dermatol Ther (Heidelb). 2022;12(7):1535–50.
Google Scholar
Hu S, Ding Q, Zhang W, Kang M, Ma J, Zhao L. Gut microbial beta-glucuronidase: a vital regulator in female Estrogen metabolism. Gut Microbes. 2023;15(1):2236749.
Google Scholar
Chapadgaonkar SS, Bajpai SS, Godbole MS. Gut Microbiome influences incidence and outcomes of breast cancer by regulating levels and activity of steroid hormones in women. Cancer Rep. 2023;6(11):e1847.
Chapadgaonkar SS, Bajpai SS, Godbole MS. Gut Microbiome influences incidence and outcomes of breast cancer by regulating levels and activity of steroid hormones in women. Cancer Rep (Hoboken). 2023;6(11):e1847.
Google Scholar
Haslam DE, Li J, Dillon ST, et al. Stability and reproducibility of proteomic profiles in epidemiological studies: comparing the Olink and SOMAscan platforms. Proteomics. 2022;22:13–4.
Smith JG, Gerszten RE. Emerging Affinity-Based proteomic technologies for Large-Scale plasma profiling in cardiovascular disease. Circulation. 2017;135(17):1651–64.
Google Scholar
Candia J, Cheung F, Kotliarov Y, et al. Assessment of variability in the SOMAscan assay. Sci Rep. 2017;7(1):14248.
Google Scholar