The detailed methods of Cell Lines and Cell Culture, Quantitative Real-Time PCR (RT-qPCR), Cell proliferation, migration assay, Cell survival assay, Cell apoptosis analysis, Identification of Prognosis-Related Genes in Gastric Cancer, Drug Sensitivity Analysis, Stability assay, Cellular Uptake, In vitro Transfection Efficiency are described in Supplementary Experimental Procedures.
Tissue specimens
A total of 40 pairs of fresh GC tissues and adjacent normal tissues were obtained under the supervision of gastroenterologists. These samples were maintained at − 80 °C to be used in subsequent RT-qPCR and Western blotting analyses, or fixed in 10% formalin and embedded in paraffin for immunohistochemistry (IHC). Additionally, The Pathology Department at the First Affiliated Hospital of Nanchang University provided 210 paraffin-embedded GC tissue samples. All the samples originated from patients diagnosed with GC who underwent surgical procedures between January 2017 and January 2019, none of whom had received any prior treatment. The Ethics Committee of the First Affiliated Hospital of Nanchang University approved the study, and all participants provided informed consent (Approval Number: (2025) CDYFYYLK (04–030)).
siRNA, shRNA, plasmid construction, and cell transfection
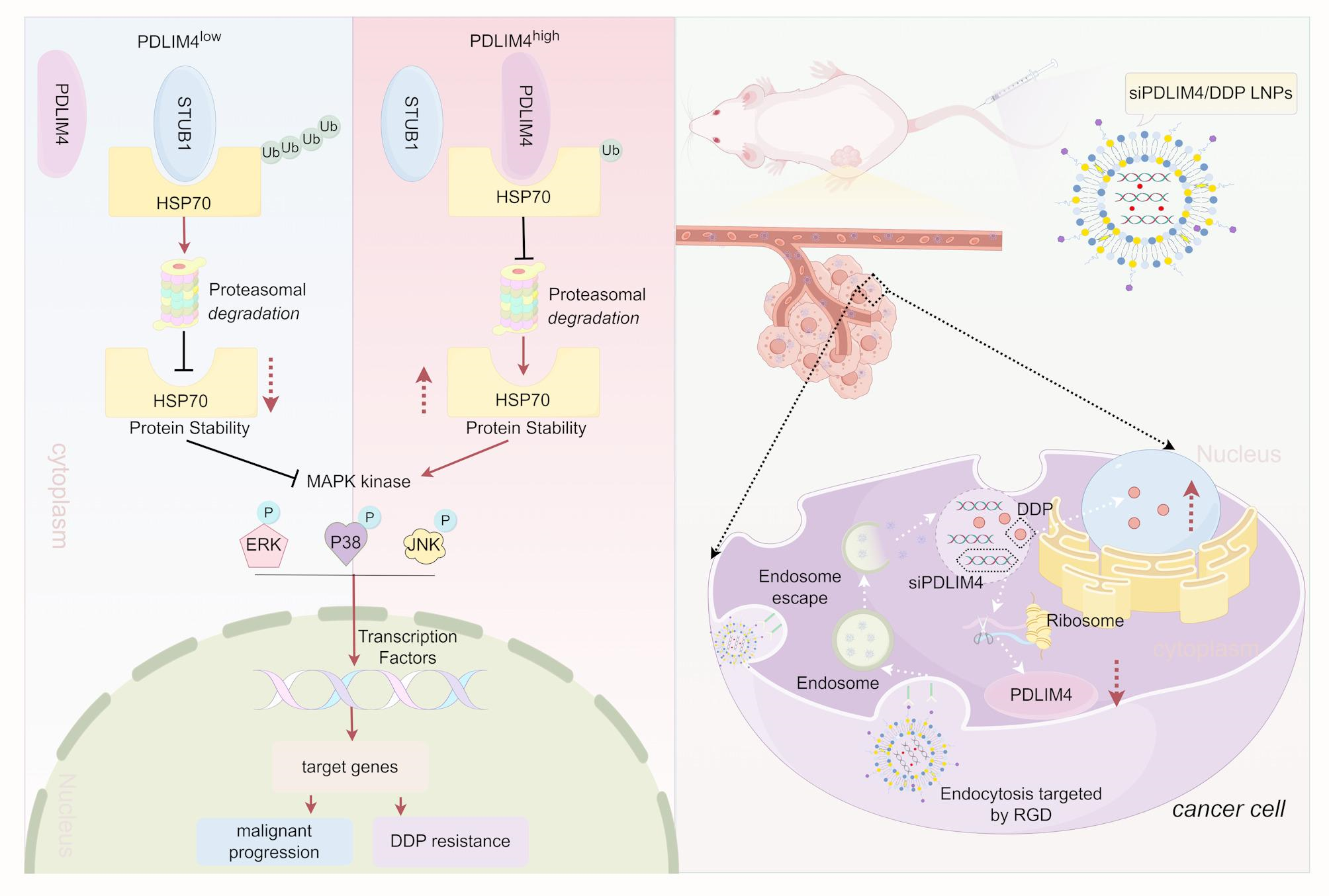
General Biol (Anhui) Co., Ltd (Anhui, China) synthesized the siRNA and shRNA primers, with their sequences listed in Table S1. A segment of SFB, whose sequence is also listed in Table S1, was fused to the N-terminus of PDLIM4 to facilitate binding with S-Protein Agarose and Flag antibody. This construct was inserted into the PCDH-CMV-MCS-EF1-copGFP-T2A-Puro vector, resulting in the SFB-PDLIM4 construct. Variants of SFB-PDLIM4, including SFB-PDLIM4 (330AA), SFB-PDLIM4 (1-88AA), SFB-PDLIM4 (1-260AA), SFB-PDLIM4 (80-260AA), SFB-PDLIM4 (80-330AA), and SFB-PDLIM4 (250-330AA) were procured from PPL (Public Protein/Plasmid Library, China). The HSP70 variants, HSP70-ΔN, HSP70-ΔC, and HSP70-ΔABD were procured from LuoRui Biotech (Jiangsu, China). For siRNA and plasmid transfections, Lipo3000 (Thermo Fisher Scientific, USA) and jet PRIME® (Polyplus, France) were employed, respectively. To generate stable cell lines with either knockdown or overexpression of PDLIM4, we utilized psPAX2, pMD2.G, and the target plasmid for lentiviral packaging. The resulting viral supernatant was then used to infect MKN45 and HGC27 cells.
Western blotting
Total Proteins were split by SDS-PAGE, afterwards shifted to 0.45 μm nitrocellulose membranes (Millipore, USA). The membranes were incubated overnight with the primary antibody following 2 h block using 5% milk. Membranes were exposed to a secondary antibody the next day. Image acquisition was conducted using the UVP/ChemStudio Imaging System (Analytik Jena AG, Germany). The antibodies used included PDLIM4 (Abcam, Cat# ab251701), HSP70 (Proteintech, Cat# 10995–1-AP), STUB1 (Abmart, Cat# PHM9480), β-actin (ABclonal, Cat# AC026), GAPDH (Proteintech, Cat# 81640–5-RR), MYC (Proteintech, Cat# 16286–1-AP), Ub (Proteintech, Cat# 10201– 2-AP), HA (ABclonal, Cat# AE008), HA (Proteintech, Cat# 51064–2-AP), DYKDDDDK (Proteintech, Cat# 20543–1-AP), DYKDDDDK (Proteintech, Cat# 66008–4-Ig), P21 (ABclonal, Cat# A19094), P27 (ABclonal, Cat# A19095), bcl2 (Servicebio, Cat# GB154380), Bax (Servicebio, Cat# GB11690), E-cadherin (Proteintech, Cat# 20874–1-AP), MMP2 (Wanleibio, Cat# WL03224), MMP3 (Proteintech, Cat# 66338–1-Ig), MMP9 (Huabio, Cat# ET174-69), Vimentin (Huabio, Cat# ET1610-39), ERK (Proteintech, Cat# 11257–1-AP), P38 (Proteintech, Cat# 66234–1-Ig), JNK (Proteintech, Cat# 66210–1-Ig), p-ERK (Cohesion, Cat# CQA8093), p-P38 (Proteintech, Cat# 28796–1-AP), p-JNK1/2/3 (Cohesion, Cat# CQA8172), Integrin Alpha V + Beta3 (absin, Cat# abs122318), Integrin Alpha V + Beta3 (LMAI, Cat# LM-1310R), Rabbit IgG (Proteintech, Cat# 30000–0-AP) and Mouse IgG (Proteintech, Cat# B900620).
Immunohistochemistry (IHC)
The paraffin-embedded sections underwent deparaffinization and rehydration, antigen retrieval was performed with EDTA (pH 9.0). Subsequently, they were immersed in a 3% hydrogen peroxide solution for 15 min. Sections were incubated overnight at 4 °C with primary antibodies after 2 h block with 5% milk, including PDLIM4 (Abmart, Cat# PC6093M), HSP70 (Proteintech, Cat# 10995–1-AP), and Ki67 (Huabio, Cat# HA721115). The next day, the sections were treated with a secondary antibody for 1 h, then stained with DAB and counterstained with hematoxylin. A scoring system was employed to evaluate the degree of staining, assessing the proportion of positive cells using the following scale: 0 (0%), 1 (1–24% positive cells), 2 (25–49% positive cells), 3 (50–74% positive cells), and 4 (≥ 75% positive cells). The intensity of staining was rated as 0 for negative, 1 for weak, 2 for moderate, and 3 for strong. The immune reactivity score was obtained by multiplying the degree score with the intensity score, with the median value used to differentiate between low and high expression.
Immunofluorescence (IF)
MKN45 and HGC27 cells, transfected with SFB-PDLIM4 and HSP70-HA, were fixed using 4% paraformaldehyde for a duration of 30 min. Following this, 0.2% Triton X-100 was used to permeabilize the cells for 15 min. Following permeabilization, the cells were blocked with 5% goat serum for 1 h. Primary antibodies were used to incubate the cells overnight at 4 °C, and the next day, secondary antibodies were applied for 1 h. Imaging was conducted utilizing a ZEISS confocal laser scanning microscope (Germany).
Co-immunoprecipitation
The designated plasmids were used to transfect the cells, which were then lysed using IP lysis buffer (Servicebio, China) containing 50 × Cocktail (Servicebio, China). The cell lysates underwent incubation at 4 °C overnight for co-immunoprecipitation using specified antibody in conjunction with S-Protein Agarose (Millipore, USA) or Protein A/G Plus Sepharose 4FF Affinity Chromatography Resin (Sangon Biotech, China). The beads were washed with TBS buffer at least five times the next day. The final products were boiled in 2 × SDS-PAGE loading buffer (Solarbio, China) at 100 °C for 10 min. The immunoprecipitated samples were analyzed via western blotting.
Proximity ligation assay (PLA)
The PLA was performed using a NaveniFlex Cell Red kit (Navinci, Sweden). On the first day, gastric cancer cells were seeded onto slides, and on the second day, they were fixed and permeabilized. After blocking non-specific binding with Naveni Block, the samples were incubated overnight with primary antibodies (rabbit anti-PDLIM4 and mouse anti-HSP70 (Proteintech, Cat#66183–1-Ig) or mouse anti-IgG). Navenibody were applied and incubated after 1 × TBST washing, with subsequent ligation and amplification steps. Finally, we cleaned the slides and performed nuclear staining using DAPI (Beyotime, China), and took photos using a ZEISS confocal laser scanning microscope (Germany).
Mouse xenograft model
The animal research received approval from the Ethics Committee of the First Affiliated Hospital of Nanchang University (Ethics Number: CDYFY-IACUC-202309QR019). Hangzhou Ziyuan Laboratory Animal Technology Co., Ltd. supplied female BALB/c nude mice that were four weeks old. For the mouse xenograft tumor assay, nude mice received a subcutaneous injection of about 5 million cells, and the tumor volume was measured every three days. The calculated using the formula: volume = length × width^2 × 0.5. The mice were euthanized when the tumors grew to a size of 15 mm in diameter, and the tumors were either embedded in paraffin or kept at −80 °C.
Synthesis of siPDLIM4 LNPs, DDP LNPs and siPDLIM4/DDP LNPs
Lipid nanoparticles (LNPs) were created by dissolving MC3, DSPC, DMG-PEG2000, cholesterol, and DSPE-PEG-RGD in ethanol with a molar ratio of 50:10:1.5:38.5:1. Subsequently, the lipid mixture and siRNA were combined at a nitrogen-to-phosphate (N/P) ratio of 6, which prepare LNPs encapsulating siPDLIM4 or/and DDP using a microfluidic cartridge.
Characterization of lipid nanoparticles
The size and zeta potential of the nanoparticles were measured using the Zetasizer Nano ZS90 (Malvern Instruments, UK). Transmission electron microscopy (TEM, Japan) was utilized to analyze the nanoparticles’ morphological characteristics. The encapsulation efficiency of siPDLIM4 was assessed using ultraviolet–visible spectroscopy (UV–Vis), while the encapsulation efficiency of DDP was assessed using inductively coupled plasma optical emission spectrometry (ICP-OES). Firstly, we used ICP-OES: Agilent 5110 to detect the Pt content, which is converted to 1 μg/ml Pt≈1.538 μg/ml DDP. We diluted the siPDIM4/DDP LNPs 50 times and measured the average concentration of Pt is 3.786 mg/l, so the Pt concentration in the siPDIM4/DDP LNPs is 189.3 mg/l, which is equivalent to DDP concentration of 291.16 mg/l. We synthesized the siPDIM4/DDP LNPs a total of 10 ml, therefore the content of DDP is 2.9116 mg. 5 mg DDP were used in our synthetic material, the encapsulation efficiency of DDP is 2.9116 mg/5 mg = 58.23%. The concentration of the siPDIM4/DDP LNPs is 3 mg/ml and the volume is 10 ml, therefore, the DDP loading capacity = encapsulated DDP mass/encapsulated DDP mass + carrier mass (2.9116 mg/2.9116 mg + 30 mg = 8.85%).
Cisplatin release profile from LNPs
We conducted a drug release curve in PH6.0 PBS, which is a tumor-mimicking environments. We used the siPDIM4/DDP LNPs with a volume of 1 ml and a concentration of 3 mg/ml for drug release. Then we put it into a dialysis bag, the dialysis solution was 30 ml PBS buffer with pH 6.0. Later, we took out 1 ml dialysis solution at regular intervals and measured the content of Pt released by ICP-OES: Agilent 5110, and converted it to DDP content. The cumulative release = release DDP amount/total DDP amount.
Pharmacokinetic assay and biodistribution of SIPDIM4/DDP LNPS in mice
Healthy female mice were assigned into 3 groups randomly (n = 3), with every group receiving a tail vein injection of either naked siPDLIM4, or siPDLIM4 LNPs, siPDLIM4/DDP LNPs, each at a dose of 1 nmol siRNA. The orbital vein was used to collect blood samples using heparinized tubes at predetermined intervals. The fluorescence intensity of CY5-labeled siPDLIM4 in the blood was calculated using a microplate reader to assess pharmacokinetics. For the biodistribution study, healthy female BALB/c nude mice with subcutaneous tumors were randomly divided into two groups (n = 16), with each group receiving an intravenous injection of either naked Cy5-siPDLIM4 or Cy5-siPDLIM4/DDP LNPs. After 1, 6, 24 h, the fluorescence intensity of CY5-siPDLIM4 in major organs and tumors was measured using the small animal CT/live imaging all-in-one machine (Milabs B.V.).
Anti-tumor efficacy and toxicity of siPDLIM4 LNPs, DDP LNPs and siPDLIM4/DDP LNPs
In the mouse xenograft tumor assay, nude mice received subcutaneous injections of approximately 5 million MKN45 cells. Then, on the sixth day the mice of xenograft tumors were divided into five groups (n = 6) through random allocation. On the seventh day, the mice received injections via the tail vein (This injection method refers to relevant literatures [26, 29, 30, 33, 34]) with either PBS, LNPs, siPDLIM4 LNPs, DDP LNPs, or siPDLIM4/DDP LNPs with 1 nmol siRNA or/and DDP dosage of 2 mg/kg, administered every three days for a total of six cycles, which primarily informed by existing literatures [35,36,37]. On the 25th day, the mice were executed, and their major organs and tumors were harvested and preserved in 4% paraformaldehyde for IHC and HE staining. Servicebio (Wuhan, China) conducted an analysis of blood biochemical parameters such as alanine aminotransferase (ALT), aspartate aminotransferase (AST), urea, and creatinine (Crea).
Data sources
The Cancer Genome Atlas (TCGA) database (http://cancergenome.nih.gov/) provided us with clinical information and RNA-seq transcriptomic data on GC. Additionally, four datasets—GSE66229, GSE122401, GSE179252, and GSE184336 were derived from the Gene Expression Omnibus (GEO) database (http://www.ncbi.nlm.nih.gov/geo/). The GDSC2 drug sensitivity data were accessed from the Genomics of Drug Sensitivity in Cancer (GDSC) database via GitHub (https://github.com/maese005/oncoPredict/tree/main/vignettes). Samples lacking complete clinical information were deemed ineligible and subsequently excluded from the study. Following the preprocessing of raw data, which encompassed sample deduplication, data normalization, probe annotation, gene filtering, and clinical grouping. To analyze differential gene expression between tumor and normal tissues, we used the Lima package in R.
Statistical analysis
We used R and GraphPad Prism 9.0 for performing statistical analyses. Results are displayed as the mean ± standard deviation based on three different experiments. For differences between two groups, a t-test was utilized, whereas two-way or one-way ANOVA was employed for comparisons among multiple groups. Spearman’s rank order correlation was used to perform the correlation analysis. A P-value below 0.05 was regarded as a sign of statistical significance.