Despite being banned in Pakistan, Dhurandhar has shattered records with massive box-office collections and unprecedented illegal downloads.
Author: admin
-

Banned in Pakistan, ‘Dhurandhar’ breaks records with 2 million illegal downloads in just 12 days
Ranveer Singh and Sara Arjun in Dhurandhar movie Continue Reading

ICC T20 World Cup 2026: India's probable squad for title defence – Deccan Herald
- ICC T20 World Cup 2026: India’s probable squad for title defence Deccan Herald
- Gill misses out on India’s T20 World Cup 2026 squad ICC
- Gill left out of India’s T20 World Cup squad; Rinku, Kishan make comebacks ESPNcricinfo
- Shubman Gill dropped by…
Continue Reading

We caught up with one of the kids who sang on ‘A Charlie Brown Christmas.’ He’s 72 now
Dave Willat was just 11 years old, wearing cutoffs and a T-shirt on a warm September evening in 1965, when he showed up for what he thought would be a routine choir practice at his church in San Rafael,…
Continue Reading

GI Partners to acquire Netwatch
Our cross-disciplinary team is led by Cian McCourt (Corporate and M&A) and Cormac O’Donoghue (Corporate and M&A), and includes Ailish Finnerty (Tax), Colin Rooney (Technology and Innovation), David Molloy (Debt Finance), Kevin Langford (Employment), Richard Ryan and Patrick Horan (Competition and Regulated Markets), Philip Smith (Pensions and Employee Benefits) and Simon Hannigan (Real Estate) working in close collaboration with a team from Paul, Weiss, Rifkind, Wharton & Garrison LLP.
Our full Dublin team included Graham Murtagh and Maria Colom Lawlor (Corporate and M&A), Emily Tyler and Aoibhín Ní Dhubháin (Competition and Regulated Markets), Freda McCusker (Debt Finance), Grace-Ann Meghen and Gavan McLaughlin (Employment), Michael Shovlin and Alan Harney (Pensions and Employee Benefits), Lucy Byrne (Real Estate), Nicola Cavey and Rachel Coyle (Tax), and Aoife Coll and Kerry Burns (Technology and Innovation).
The team was also supported by the work of our colleagues in Belfast, Richard Armstrong and Reuben Kane (Corporate and M&A Northern Ireland), and Chris Fullerton and Madison Bowyer (Employment Northern Ireland).
For more information on the transaction, please visit the GI Partners website.
Continue Reading

From biting flies to feathered dinosaurs, scientists reveal 70 new species
From biting fruit flies and a tiny long-nosed mouse opossum to a feathered dinosaur preserved with evidence…
Continue Reading

From biting flies to feathered dinosaurs, scientists reveal 70 new species
From biting fruit flies and a tiny long-nosed mouse opossum to a feathered dinosaur preserved with evidence…
Continue Reading
Word Circle Number 89
Unlock the Editor’s Digest for free
Roula Khalaf, Editor of the FT, selects her favourite stories in this weekly newsletter.
Download puzzle
FT.com will bring you the crossword from Monday to Saturday as well as the Weekend FT Polymath, Word…
Continue Reading
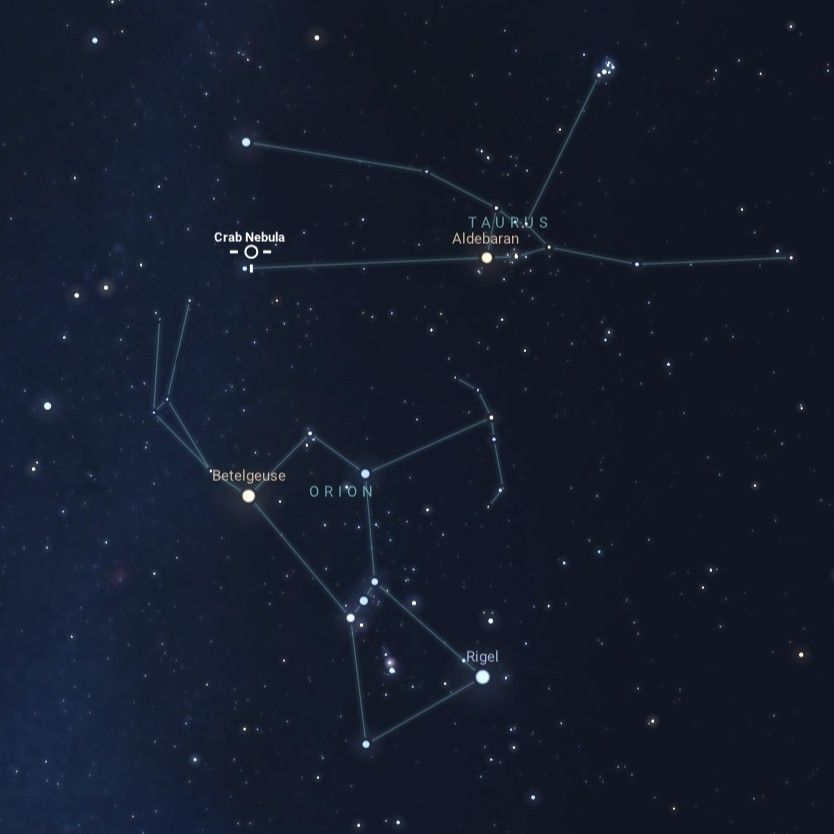
Betelgeuse and the Crab Nebula: Stellar Death and Rebirth
What happens when a star dies? In 2019, Betelgeuse dimmed in brightness, sparking speculation that it may soon explode as a supernova. While it likely won’t explode quite yet, we can preview its fate by observing the nearby Crab…
Continue Reading

Ursid meteor shower 2025: When and where to see ‘shooting stars’ on the longest night of the year
If you’re stargazing late overnight on Dec. 21-22, keep an eye out for “shooting stars” as the annual Ursid meteor shower peaks.
Although the Ursids are active from Dec. 13 through Dec. 26, the peak night coincides with the winter solstice,…
Continue Reading

‘Fire Country’ draws inspiration from firefighters who fought L.A. fires
A worried mother working miles away from her family frantically tries to get her three children out of harm’s way as an out of control fire ravages their community. Firefighters do everything they can, but strong winds are working against them…