- ESA Publishes ‘Zero Debris’ Framework Amid Effort to Deorbit Dangerous Space Junk extremetech.com
- Space trash: Orbit shows where the circular economy breaks down Bulletin of the Atomic Scientists
- Thousands of dead satellites in orbit are a big…
Category: 7. Science
-
ESA Publishes 'Zero Debris' Framework Amid Effort to Deorbit Dangerous Space Junk – extremetech.com
-

Climate engineering would alter the oceans, reshaping marine life – new study examines each method’s risks
doesn’t require adding nutrients, but some mineral forms of alkalinity, like basalts, introduce nutrients such as iron and silicate that can impact growth.
Solar radiation modification adds no nutrients but that move nutrients around.
Shifts…
Continue Reading
-
Astronomers spot mysterious 'iron bar' in well-known Ring Nebula – Reuters
- Astronomers spot mysterious ‘iron bar’ in well-known Ring Nebula Reuters
- Destroyed planet? Giant iron bar found in the Ring Nebula may be a vaporized planet Interesting Engineering
- Earth could be vaporised by the Sun as boffins make grim end of…
Continue Reading
-
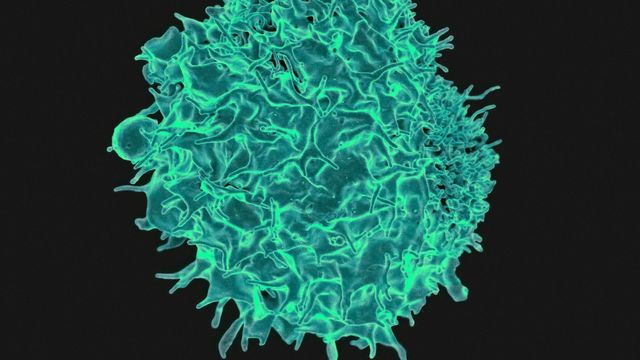
Antigens Explained: New Discovery Reshapes Immune Communication
A new discovery about how cells communicate with each other in the body’s immune system has revealed deeper insights for an international team of scientists into fundamental immune system function.
The new study, in Nature…Continue Reading
-

James Webb telescope reveals sharpest-ever look at the edge of a black hole — and it could solve a major galactic mystery
Astronomers have revealed the James Webb Space Telescope’s (JWST) sharpest-ever image of the area around a black hole. The spectacular view could help solve a decades-long mystery while reversing a long-held belief about space’s most extreme…
Continue Reading
-

The Sky Today on Monday, January 19: Where Messier started: M1
The Crab Nebula (M1) is the object that started Messier on his list of faint fuzzies. Observe it tonight in Taurus the Bull.
Looking for…
Continue Reading
-
Laser timing tech sharpens black hole radio views
by Riko Seibo
Tokyo, Japan (SPX) Jan 19, 2026
Radio telescopes capture faint radio signals from space and convert them into images of distant celestial objects, but resolving black holes sharply requires many instruments to observe…
Continue Reading
-
‘Death by a thousand cuts’: young galaxy ran out of fuel as black hole choked off supplies – Technology Org
- ‘Death by a thousand cuts’: young galaxy ran out of fuel as black hole choked off supplies Technology Org
- Measurement of the gas consumption history of a massive quiescent galaxy Nature
- ‘Death by a thousand cuts’: James Webb Space Telescope…
Continue Reading
-

Rosette Nebula, also called the Skull Nebula, 10,000 times the mass of the Sun, peaks in winter skies: How to find it
Winter is shaping up to be the ideal season for stargazers hoping to catch a glimpse of one of the night sky’s most striking sights: the Rosette Nebula. Located about 5,000 light-years from Earth, the Rosette Nebula is a vast star-forming…Continue Reading
-
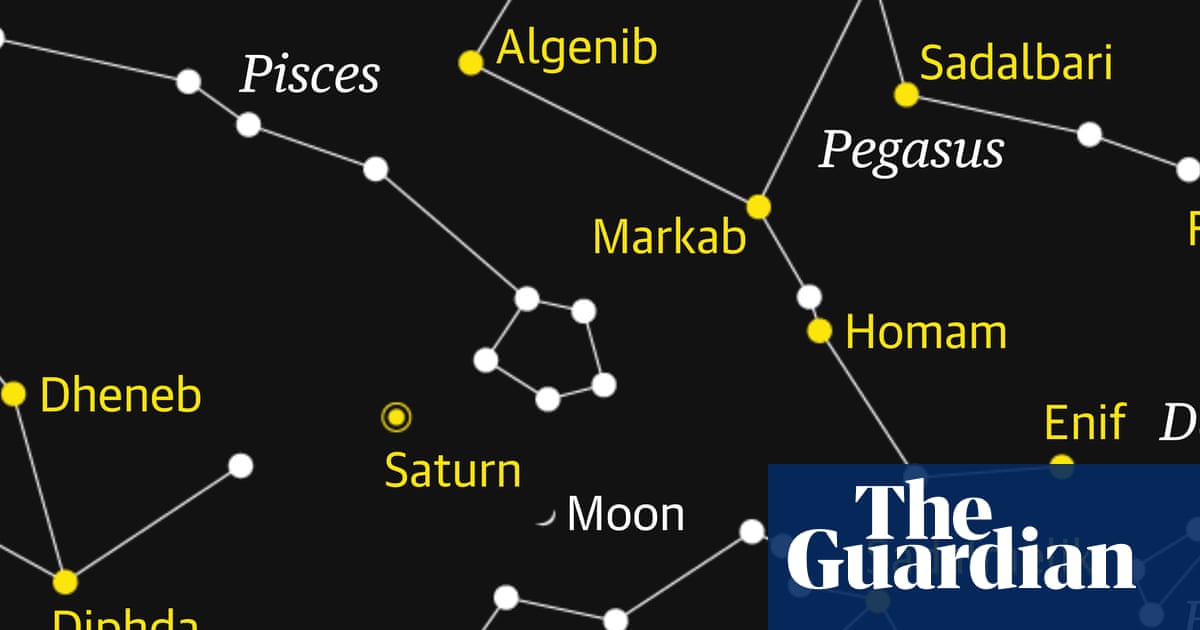
Starwatch: Watch the crescent moon pass Saturn as dusk gathers | Astronomy
A slender crescent moon slides past Saturn this week, offering a rewarding conjunction. It will be the perfect way to start your evening, a little quiet contemplation of the night sky as the evening twilight gives way to full darkness.
The chart…
Continue Reading