- A23a, once world’s largest iceberg, could disappear within weeks news.cgtn.com
- Massive iconic iceberg turns blue and is “on the verge of complete disintegration,” NASA says CBS News
- Earth from Space: The fate of a giant European Space Agency
Category: 7. Science
-

A23a, once world's largest iceberg, could disappear within weeks – news.cgtn.com
-

The World’s Longest-Running Lab Experiment Is Almost 100 Years Old : ScienceAlert
Sometimes science can be painfully slow. Data comes in dribs and drabs, truth trickles, and veracity proves viscous.
The world’s longest-running lab experiment is an ongoing work in sheer scientific patience. It has been running continuously…
Continue Reading
-

SpaceX Falcon 9 rocket launches 29 Starlink satellites to orbit from Florida
Another Starlink launch is now in the record books.
SpaceX on Sunday (Jan. 18) sent a new batch of 29 Starlink satellites (Group 6-100) into low Earth orbit. At 6:31 p.m. EDT (2331 GMT), the company launched a Falcon 9 rocket from Complex 40 at…
Continue Reading
-

Study shows why trees don’t grow faster when CO2 levels are high
Plants absorb carbon dioxide and water, use sunlight to make sugars, and release oxygen. So if the air contains more CO₂, shouldn’t forests grow faster, store more carbon, and help cool the planet?
It’s plausible and often repeated, but it…
Continue Reading
-
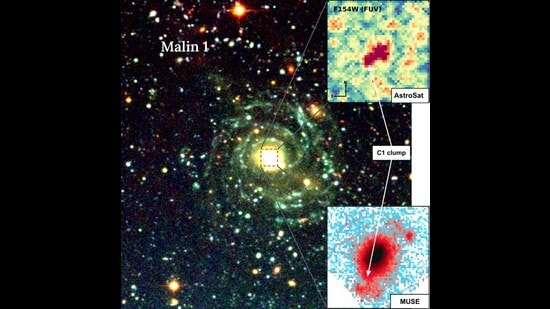
Malin 1’s secret diet: Giant galaxy found quietly cannibalising smaller neighbours
Astronomers have uncovered hidden evidence that Malin 1, the largest known low-surface-brightness galaxy in the universe, is quietly growing by swallowing smaller dwarf galaxies, a process that had remained invisible until now.
One clump, named… Continue Reading
-

Toxic gas wiped out half of all ocean life 530 million years ago
About 530 million years ago, during the early rise of complex animal life, a massive marine die-off erased roughly 45% of ocean species.
Sediment cores from the Yangtze Platform in South China point to hydrogen sulfide, a toxic gas produced in
Continue Reading
-

Ashwagandha aids recovery without blunting training stress in athletes
A controlled trial in team-sport athletes suggests Ashwagandha may help maintain hormonal balance and support recovery and power adaptations during the physiological strain of pre-season training.
Study: Ashwagandha Root Extract…
Continue Reading
-
Time-lapse of radar images shows how the Antarctic ice shelf collapses – MSN
- Time-lapse of radar images shows how the Antarctic ice shelf collapses MSN
- Watch This Glacier Race into the Sea Nautilus | Science Connected
- Experts Tracked How Fast Greenland and Antarctica’s Ice Is Moving — the Numbers Are Alarming Green…
Continue Reading
-

Earth may have a “buttery” core, described as a new state of matter
Deep beneath the planet’s surface, Earth’s core behaves in a way scientists have struggled to explain for decades, with consequences for how the planet moves and protects itself.
New evidence now suggests that the solid heart of Earth is…
Continue Reading
-

Scientists Achieve Breakthrough in Shrinking Objects to Nanoscale!
A team from MIT has reported in the journal Science that they have discovered a way to shrink objects down to 1,000 times smaller than their original size, reaching the nanometric scale. How does this so-called “implosion fabrication”…
Continue Reading