January 2026 Moon Phases(NASA/JPL-Caltech / NASA)
The first new moon of the year peaks on Sunday, as one of the last major skywatching events for January.
The new moon signals the reset of the moon’s calendar, peaking for just one…

January 2026 Moon Phases(NASA/JPL-Caltech / NASA)
The first new moon of the year peaks on Sunday, as one of the last major skywatching events for January.
The new moon signals the reset of the moon’s calendar, peaking for just one…


Nasa’s giant new moon rocket has moved to the launchpad in preparation for astronauts’ first lunar fly-around in more than half a century. The trip could blast off in February.
The 98-metre (322ft) rocket began its 1mph (1.6km/h) creep from…

NASA will soon run a high‑stakes fueling test on its Artemis II rocket, a practice run that must succeed before four astronauts can fly around the moon.
The U.S. space agency inched…

A 2003 marine heat wave in the waters around Greenland continues to impact North Atlantic ocean ecosystems decades on, with a sudden and strong increase in marine heat wave frequency persisting ever since.
Marine biologists from Germany and…

For decades, scientists have debated whether early humans truly took on the largest animals around them or merely stumbled across the leftovers.
Massive bones and stone tools often turn up side by side at ancient sites, but nature can easily mix…
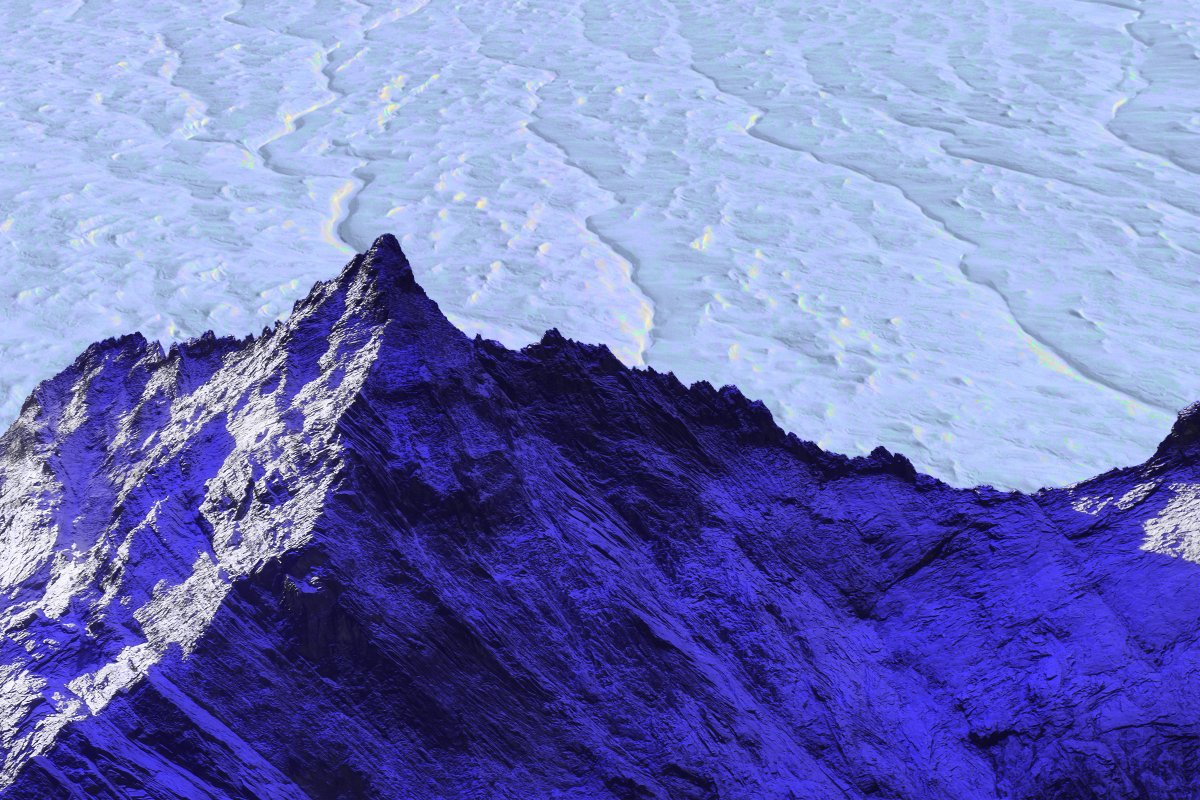
By creating a new map of Antarctica’s subterranean landscape, researchers have uncovered a vast topography of previously hidden hills,…

Far below where sunlight can reach, subtle marks in Moroccan stone suggest that life left a record in a place long assumed to be erased by time.
The finding matters because it challenges where scientists think fragile traces of early ecosystems…