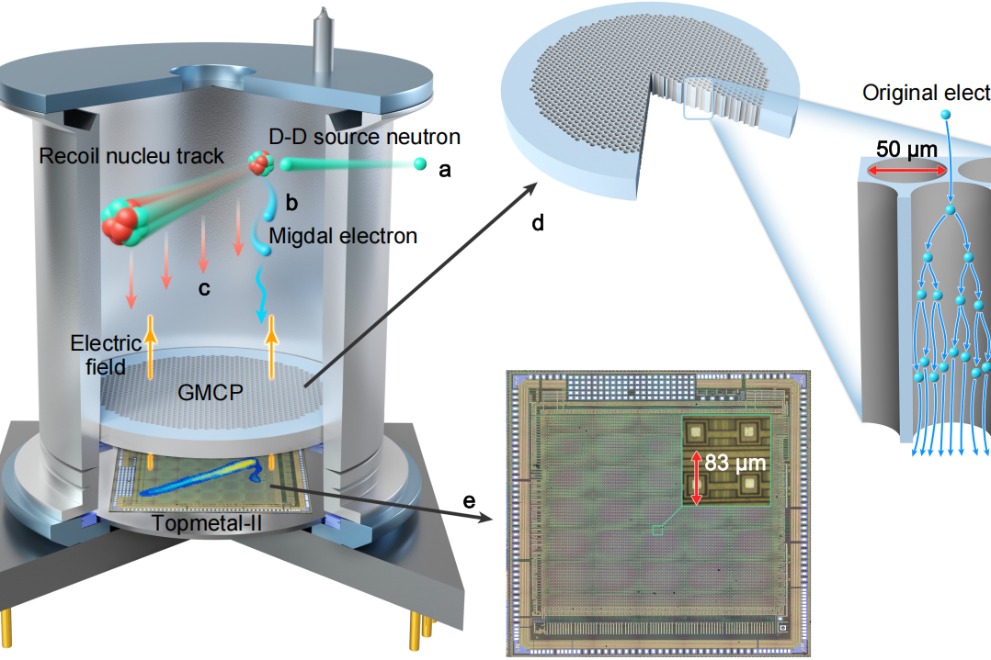
In a landmark discovery that bridges nearly a century of theoretical physics, a Chinese research team has successfully captured the first direct evidence of the Migdal effect, a breakthrough with profound implications…
Category: 7. Science
-

Tyrannosaurus rex Took Nearly Four Decades to Grow Up, New Research Shows
A comprehensive analysis of 17 fossil specimens reveals that Tyrannosaurus rex grew far more slowly than previously thought — reaching its full-grown size of eight tons around age 40 — and challenges earlier assumptions about its life…
Continue Reading
-
Habitable Worlds Observatory Living Worlds Working Group: Surface Biosignatures on Potentially Habitable Exoplanets – astrobiology.com
- Habitable Worlds Observatory Living Worlds Working Group: Surface Biosignatures on Potentially Habitable Exoplanets astrobiology.com
- How to detect signatures of alien life in exoplanet air EarthSky
- Aerial aliens: Why cloudy worlds might make…
Continue Reading
-

Beneath the ice: Satellites help map Antarctica’s subglacial surface like never before
One of the least-mapped planetary surfaces in our solar system is closer to home than you might expect: the continent of Antarctica.
While Antarctica‘s icy surface is fairly well-studied, its subglacial bedrock landscape — located up to 3…
Continue Reading
-
NASA Hustles To Replace ISS Crew After Medical Evacuation
NASA Hustles To Replace ISS Crew After Medical Evacuation | Aviation Week Network