Title: Supernova Ia Remnants with M-dwarf Surviving Companions
Authors: Kuo-Chuan Pan (潘國全), Pilar Ruiz-Lapuente, and Jonay I. González Hernández
First Author’s Institution: Department of Physics, National Tsing Hua…

Title: Supernova Ia Remnants with M-dwarf Surviving Companions
Authors: Kuo-Chuan Pan (潘國全), Pilar Ruiz-Lapuente, and Jonay I. González Hernández
First Author’s Institution: Department of Physics, National Tsing Hua…
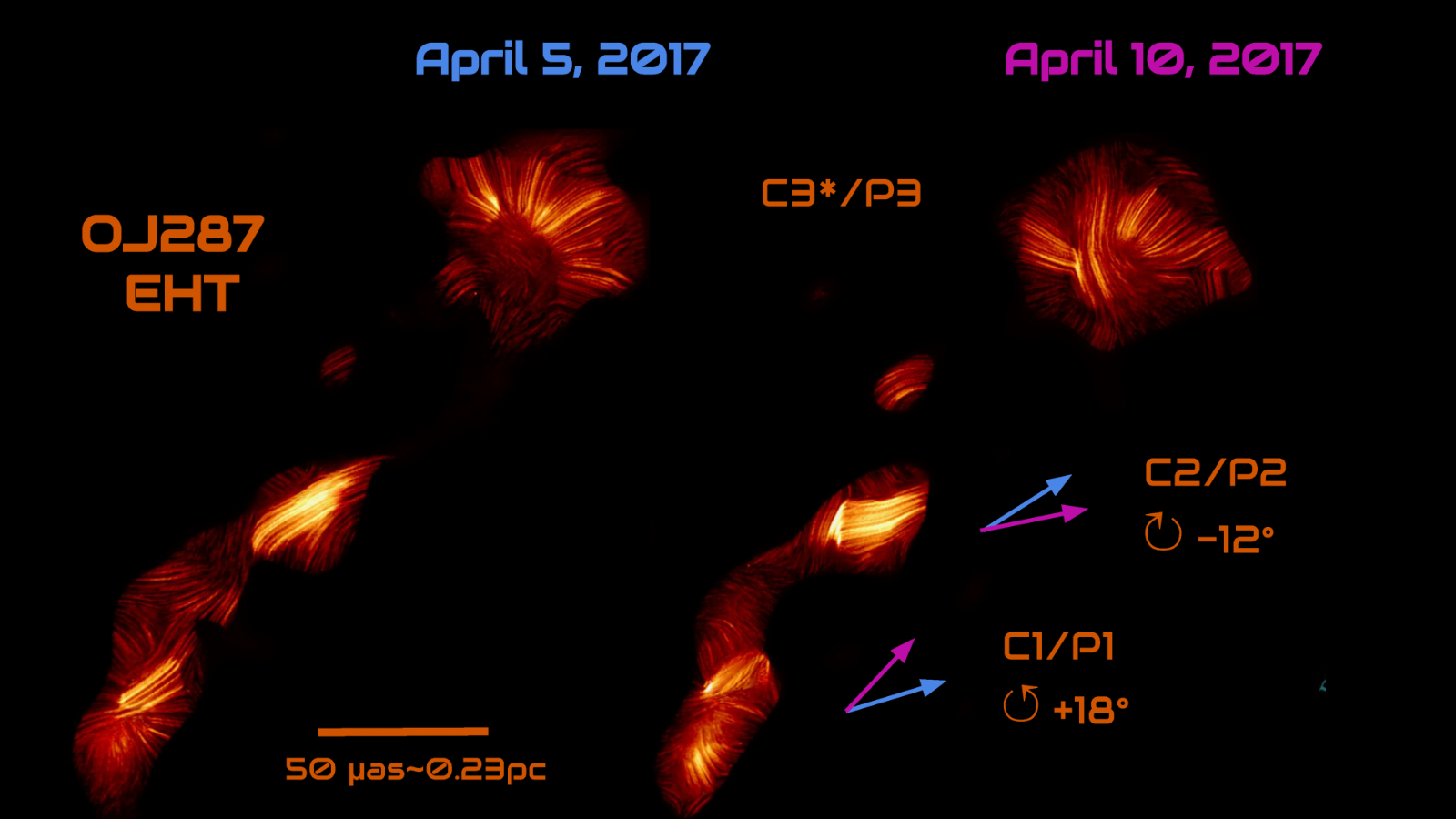
Astronomers have used the Event Horizon Telescope (EHT) to observe a violent cosmic dance between a suspected pair of supermassive black holes at the heart of a distant galaxy. The evidence for this tryst between cosmic monsters lies in the…