In this series of posts, we sit down with a few keynote speakers of the 247th AAS meeting to learn more about them and their research. You can see a full schedule of their talks here and read our other interviews here!
“We are…

In this series of posts, we sit down with a few keynote speakers of the 247th AAS meeting to learn more about them and their research. You can see a full schedule of their talks here and read our other interviews here!
“We are…

China’s first Mars rover, Zhurong, has uncovered evidence indicating that water activity on the Martian surface continued much later than scientists once believed. Researchers studying data from the rover say Mars may have experienced…

The year that iceberg A-23A first broke away from Antarctica’s Filchner Ice Shelf, Ronald Reagan was president of the United States, and the movie Top Gun was setting box office records. Forty years later, the massive tabular berg—one of…

PHOENIX — NASA says it is continuing to prepare for a possible Artemis 2 launch as soon as February, but with remarkably little publicity by the agency for the first crewed flight to the moon in more than 50 years.
In a presentation…
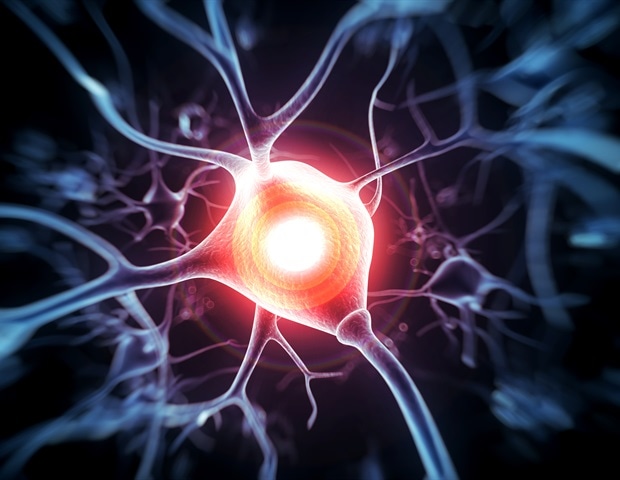
Researchers at The University of Texas MD Anderson Cancer Center have discovered that targeting a specific immune process could help improve recovery after nerve injury and reduce chronic pain.
The study, published in Proceedings…
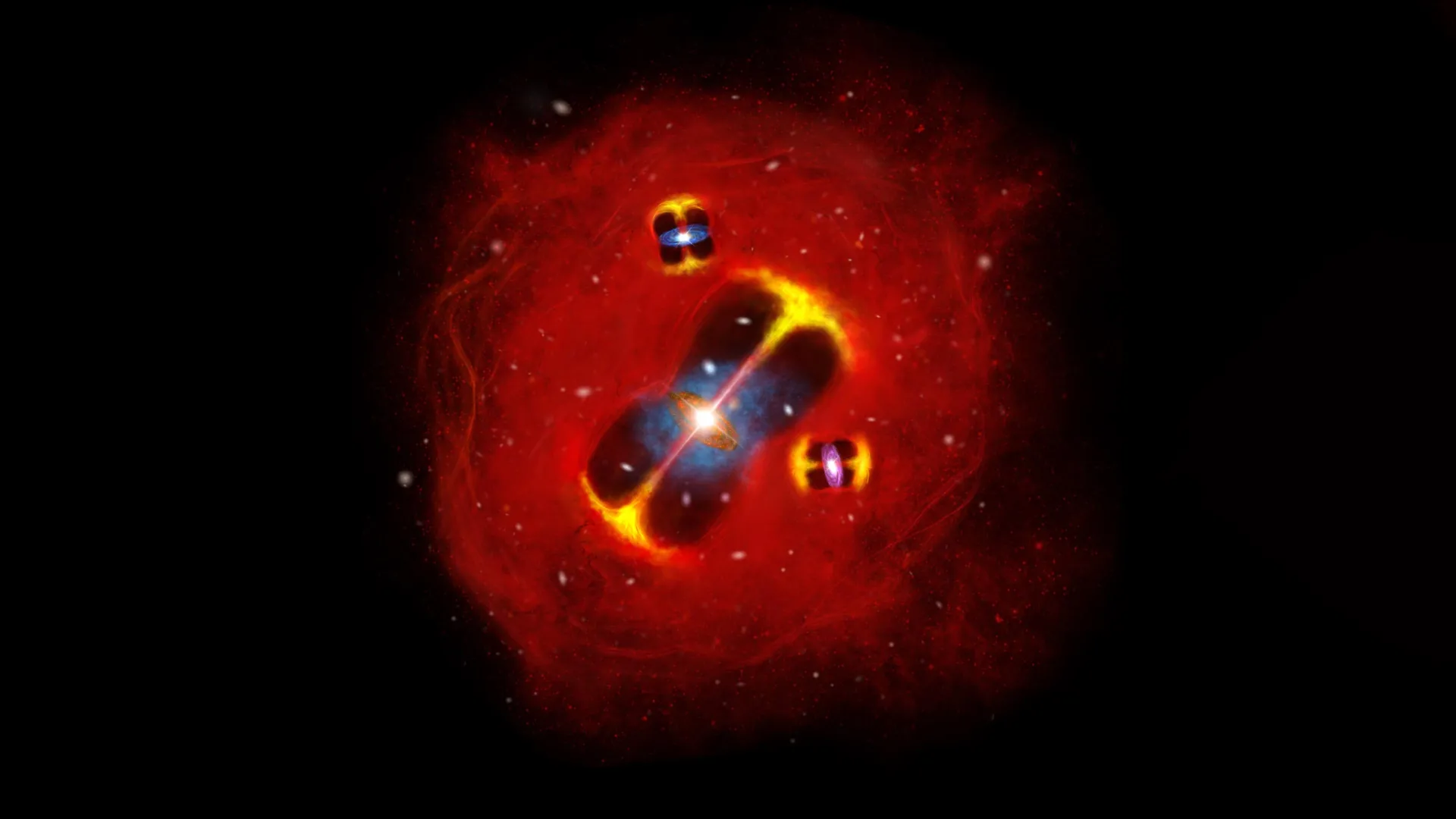
A team of astronomers from several countries, led by researchers in Canada, has identified a galaxy cluster that appears far earlier and far hotter than current science predicts. The cluster is filled with intensely hot gas and existed just 1.4…

China’s Zhurong rover has uncovered evidence that liquid water persisted on Mars far longer than scientists previously believed, according to a report by Xinhua on Wednesday citing a new peer-reviewed study.
Analysis of ground-penetrating…

by Riko Seibo
Tokyo, Japan (SPX) Jan 08, 2026
Astronomers have identified a young planetary system that links newborn giant worlds to the compact super Earths and sub Neptunes that dominate the Milky Way. V1298 Tau, a star about 20…
…
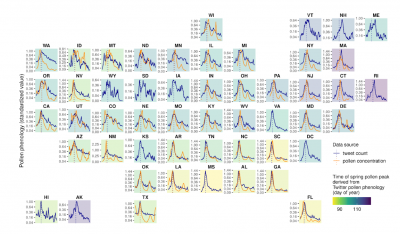
Newswise — Two things are clear from a University of Michigan analysis of nearly 200,000 Twitter posts between 2012 and 2022.
One, people are really good at identifying peak pollen season: The largest volume of…