Astronomers using the Hubble Space Telescope have identified an entirely new kind of object in the Universe.
This strange object is a starless, gas-rich cloud that’s dominated by dark matter, and is thought to be a remnant left over from the…
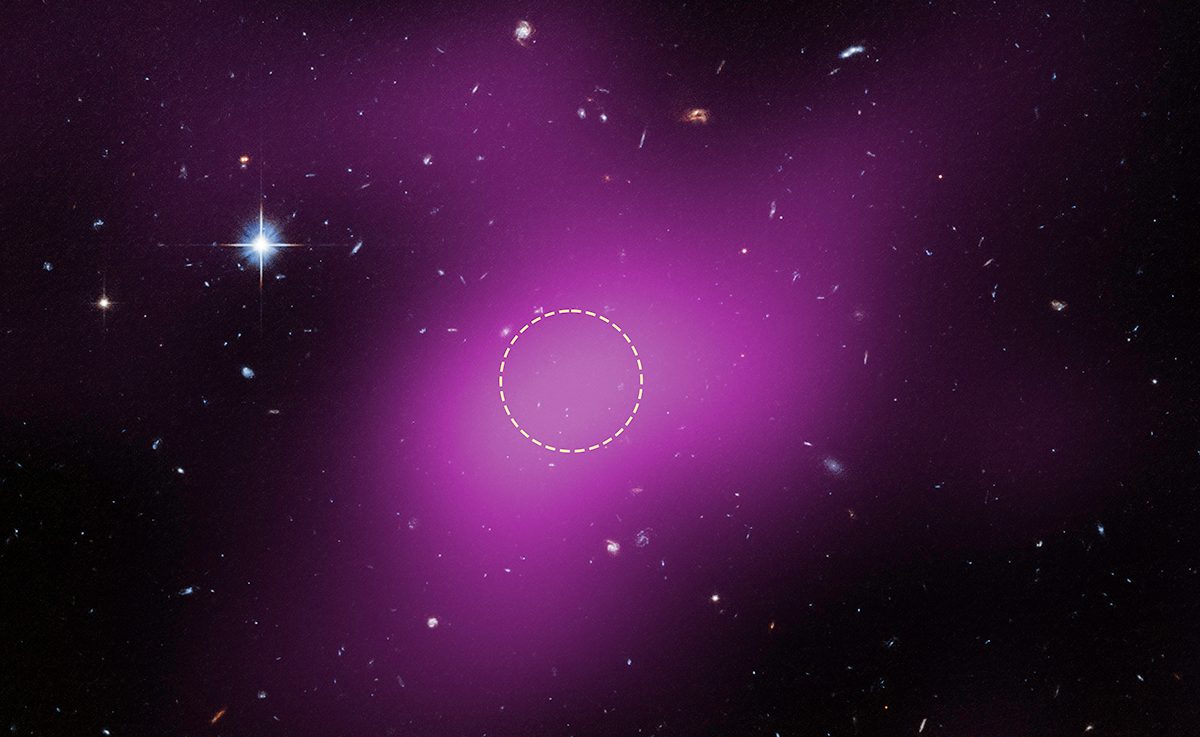
Astronomers using the Hubble Space Telescope have identified an entirely new kind of object in the Universe.
This strange object is a starless, gas-rich cloud that’s dominated by dark matter, and is thought to be a remnant left over from the…

The University of California Irvine issued a press release in which they describe identifying a DNA mechanism that enables Greenland sharks to maintain their vision over centuries.
Dorota Skowronska-Krawczyk, PhD, who is a UC Irvine associate…
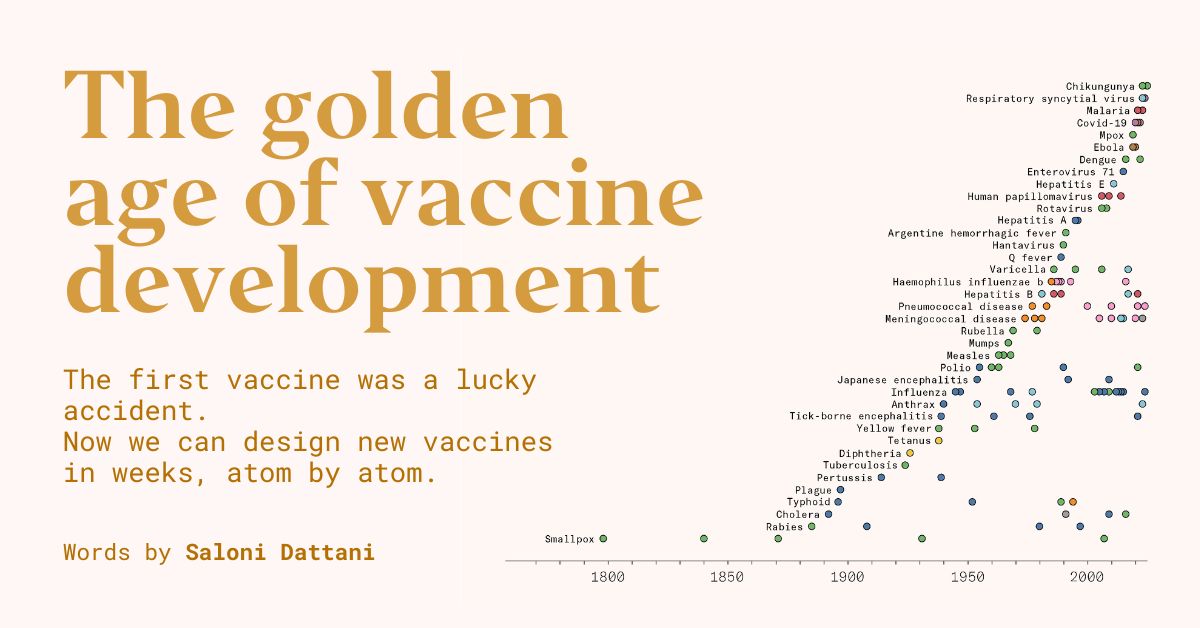
In 1796, when Edward Jenner developed the first vaccine, against the smallpox virus, no one knew what viruses were, let alone connected them to diseases.
Many believed Jenner’s vaccine worked because it depleted the body of the specific…

A cosmic first has been achieved. For the first time, scientists have used NASA’s Imaging X-ray Polarization Explorer (IXPE) to look into the turbulent heart of a white dwarf star. Their target is EX Hydrae, a restless stellar remnant in the

The field of theoretical neuroscience, which uses computational models to predict neural activity in the brain, is relatively new. Twenty years ago, there were…

ALMA Studies Supersized Protoplanetary Disk
New observations with the Atacama Large Millimeter/submillimeter Array (ALMA) have provided astronomers with an unprecedented view into the…
If you want a job done…

Ever since it was first spotted hurtling through the solar system, Harvard astronomer Avi Loeb has maintained that there’s a chance mysterious interstellar object 3I/ATLAS is a technological object sent by an extraterrestrial civilization.
Continue Reading