Physicists like Sean Carroll argue not only that quantum mechanics is not only a valuable way of interpreting the world, but actually describes reality, and that the central equation of quantum mechanics – the wave function – describes…
Category: 7. Science
-
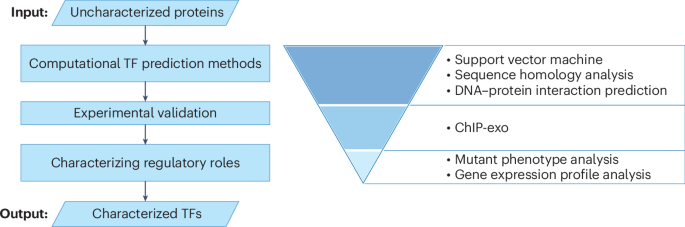
Approaches for accelerating microbial gene function discovery using artificial intelligence
Hutchison, C. A. I. et al. Design and synthesis of a minimal bacterial genome. Science 351, aad6253 (2016).
Google Scholar
Baek, M. et al. Accurate prediction of protein structures…
Continue Reading
-

China's Mars rover discovers longer water existence on red planet – news.cgtn.com
- China’s Mars rover discovers longer water existence on red planet news.cgtn.com
- Mars: Once Habitable, or Just Deceptively Earth-Like? vocal.media
- Scientists just found the best places to look for ancient life on Mars ScienceDaily
- What are the…
Continue Reading
-
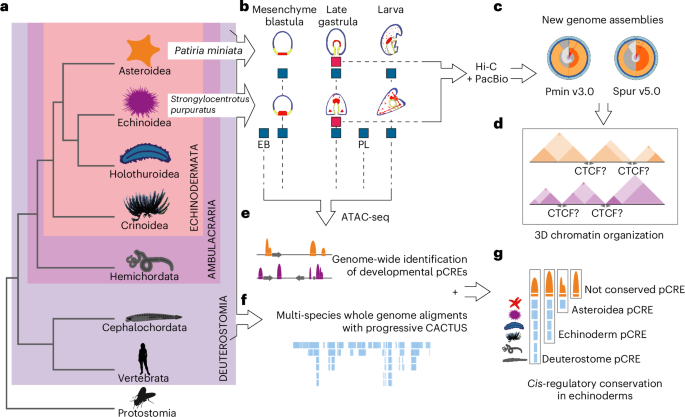
Deep conservation of cis-regulatory elements and chromatin organization in echinoderms uncover ancestral regulatory features of animal genomes
Sebé-Pedrós, A. et al. The dynamic regulatory genome of capsaspora and the origin of animal multicellularity. Cell 165, 1224–1237 (2016).
Google Scholar
Continue Reading
-
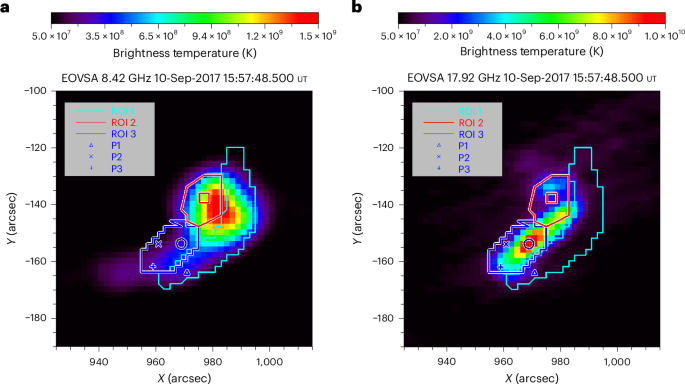
Megaelectronvolt-peaked electrons in a coronal source of a solar flare
Yokoyama, T. & Shibata, K. Magnetic reconnection as the origin of X-ray jets and Hα surges on the Sun. Nature 375, 42–44 (1995).
Google Scholar
Fleishman, G. D. et al. Decay of the…
Continue Reading
-
An intelligent hybrid deep learning-machine learning model for monthly groundwater level prediction
Tao, H. et al. Groundwater level prediction using machine learning models: A comprehensive review. Neurocomputing 489, 271–308 (2022).
Sun, J., Hu, L., Li, D., Sun, K. & Yang, Z. Data-driven…
Continue Reading
-

What space archaeology reveals about human culture after 25 years aboard the International Space Station
The International Space Station is one of the most remarkable achievements of the modern age. It is the largest, most complex, most expensive, and most durable spacecraft ever built.
Its first modules were launched in 1998. The…
Continue Reading
-

Accessible and engaging, ‘Strata’ looks back, way back, at air, ice, mud and heat
“Strata: Stories from Deep Time” by Laura Poppick. W. W. Norton & Company. $29.99 It’s a challenge for the human brain to reckon with deep time. Visits to the Great Pyramids or Stonehenge can jog us out of present-tense bias and…
Continue Reading
-

The secret world of animal sleep
Every animal with a brain needs sleep — and even a few without a brain do, too. Humans sleep, birds sleep, whales sleep and even jellyfish sleep. Sleep is universal “even though it’s actually very risky,” said Paul-Antoine Libourel, a…
Continue Reading
-

We study glaciers. ‘Artificial glaciers’ and other tech may halt their total collapse | Brent Minchew and Colin Meyer
Sea levels are rising faster than at any point in human history, and for every foot that waters rise, 100 million people lose their homes. At current projections, that means about 300 million people will be forced to move in the decades to come,…
Continue Reading