While peering into some of the oldest regions of the known universe — dating back to just 1.4 billion years after the Big Bang — an international team of astronomers made a puzzling discovery.
As detailed in a paper published the journal…
While peering into some of the oldest regions of the known universe — dating back to just 1.4 billion years after the Big Bang — an international team of astronomers made a puzzling discovery.
As detailed in a paper published the journal…

The giant planet Jupiter has nearly 100 known moons, yet none have captured the interest and imagination of astronomers and space scientists quite like Europa, an ice-shrouded world that is thought to possess a vast ocean of liquid salt…
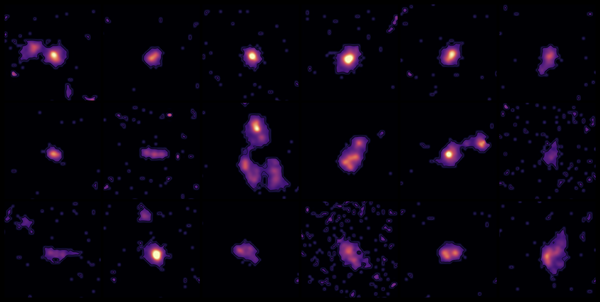
Astronomers have captured the most detailed look yet at faraway galaxies at the peak of their youth, an active time when the adolescent galaxies were fervently producing new stars. The observations focused on 18 galaxies located 12.5 billion…

Newswise — Using NASA’s James Webb Space Telescope, astronomers have spotted two rare kinds of dust in the dwarf galaxy Sextans A, one of the most chemically primitive galaxies near the Milky Way. The finding of…

Jan. 6, 2026
Contact: Eric Stann, StannE@missouri.edu
Photo by Abbie Lankitus
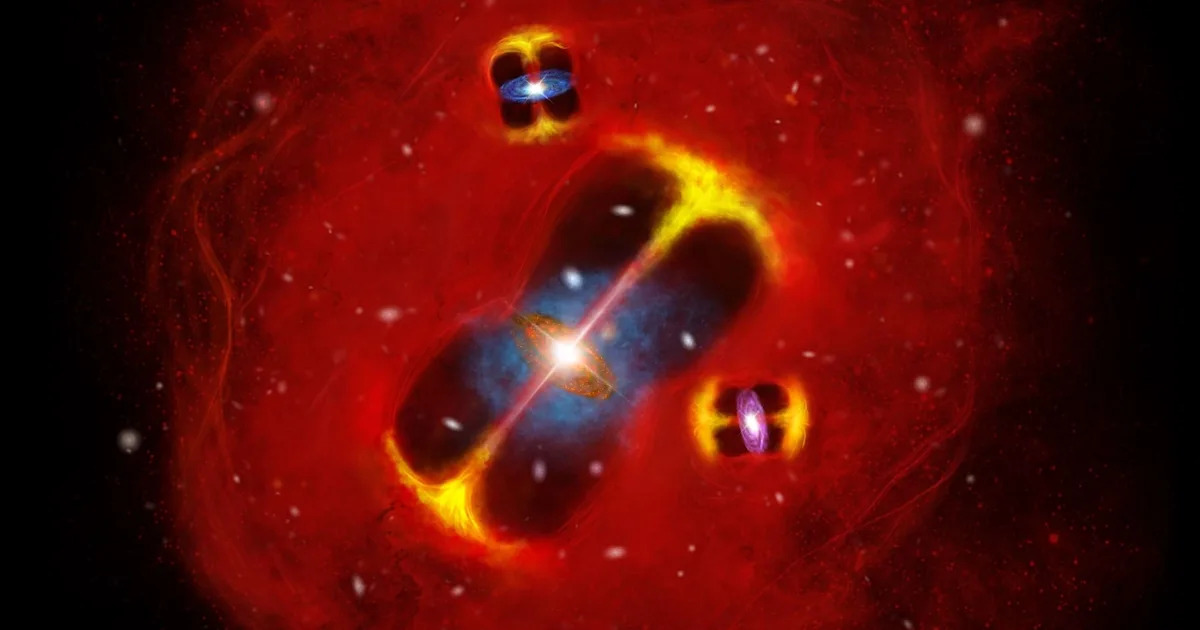
Scientists at the University of Missouri have identified a small group of unusual objects in the early universe. Using…
Phoenix, AZ (January 6, 2026)— Using data from NASA’s James Webb Space Telescope, astronomers from the Center for Astrophysics | Harvard & Smithsonian (CfA) have revealed the universe’s most mysterious distant objects, known as little red dots,…

The oldest fossilised remains of complex animals appear suddenly in the fossil record, and as if from nowhere, in rocks that are 538 million years old.
The very oldest of these are simple fossilised marks (called Treptichnus) made by

After combing through NASA’s James Webb Space Telescope’s archive of sweeping extragalactic cosmic fields, a small team of astronomers at the University of Missouri says they have identified a sample of galaxies that have a previously…

Using NASA’s James Webb Space Telescope, astronomers have spotted two rare kinds of dust in the dwarf galaxy Sextans A, one of the most chemically primitive galaxies near the Milky Way. The finding of metallic iron dust and silicon carbide…
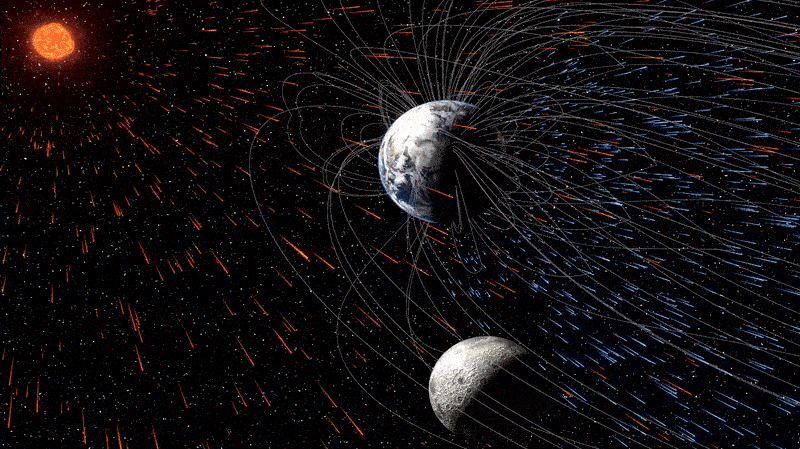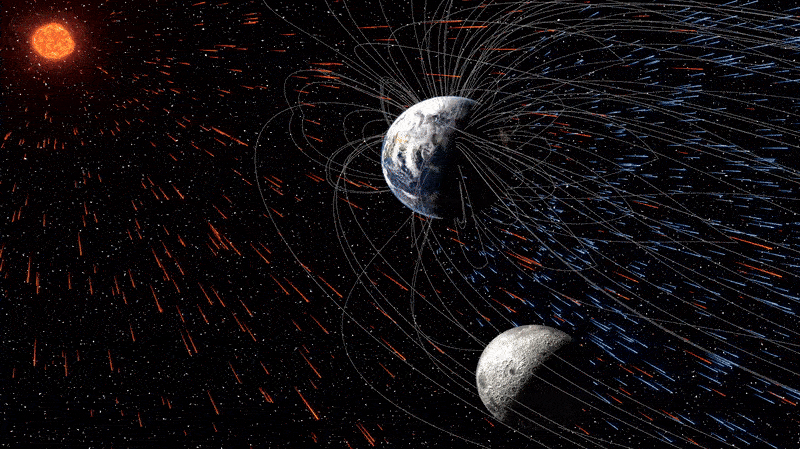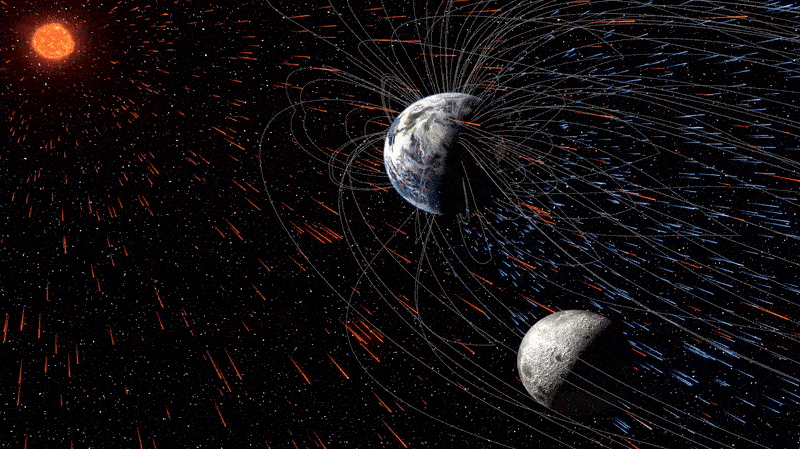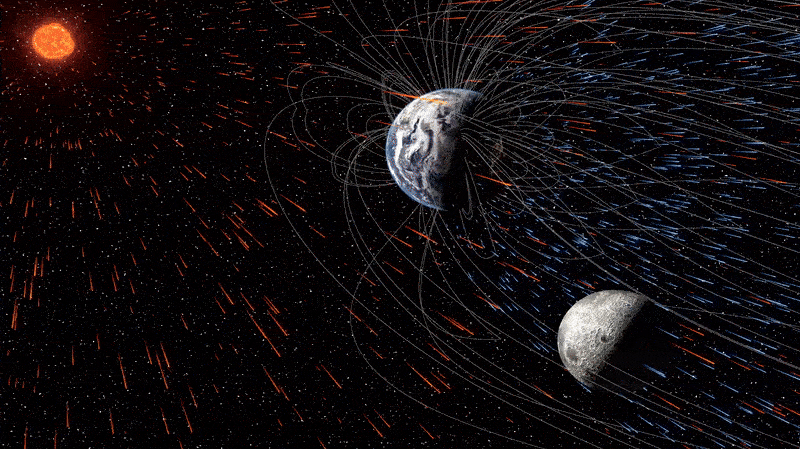
The moon is quietly absorbing tiny fragments of Earth’s atmosphere — and has been doing so for billions of years, a new study reveals. This surprising case of cosmic cannibalism is thanks to supercharged solar winds and, more importantly, our…