Recently, the James Webb Space Telescope (JWST) took a stunning image of the star cluster known as Westerlund 2, located in a stellar nursery called Gum 29 found within the Carina Nebula. The cluster is 6-to-13 light-years across and has some…
Category: 7. Science
-
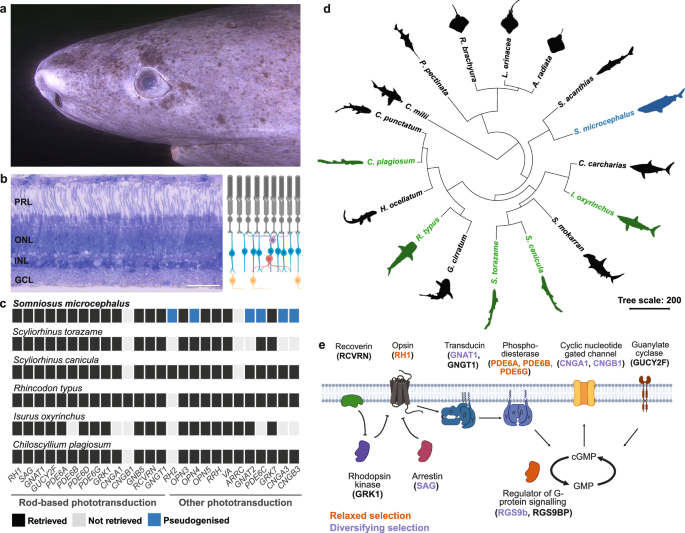
The visual system of the longest-living vertebrate, the Greenland shark
Bloch, M.E. and J.G. Schneider. M.E. Blochii, Systema ichthyologiae iconibus CX illustratum. Vol. [Atlas]. Berolini: Sumtibus auctoris impressum et Bibliopolio Sanderiano commissum. (Bavarian State Library, 1801).
Nielsen, J. et al. Eye lens…
Continue Reading
-

Scientific publisher agreements expand open-access research
UNIVERSITY PARK, Pa. — Penn State University Libraries will enter into new open-access publishing agreements with two major scientific publishers starting in 2026. These contracts ensure that Penn State peer-reviewed research published through…
Continue Reading
-
Why do some brains adapt to change faster than others? New study offers clues
Why are some people better at switching tasks than others?
A new study by researchers at Rutgers University showed that the brain’s wiring and timing together help explain why some people switch between mental tasks more efficiently, and why…
Continue Reading
-

The Ambitious Plan to Spot Habitable Moons Around Giant Planets
So far, humanity has yet to find its first “exomoon” – a Moon orbiting a planet outside of the solar system. But that hasn’t been for lack of trying. According to a new paper by Thomas Winterhalder of the European Southern…
Continue Reading
-

January full moon wows skywatchers with a striking ‘Wolf Supermoon’ (photos)
The January full moon soared through the winter sky on Jan. 3, putting on an awe-inspiring display as it flooded the night with reflected sunlight to kick off a new year of spectacular lunar milestones. Read on to see jaw-dropping images of the…
Continue Reading
-

Scientists found the most heat-tolerant plant on Earth hiding in plain sight
In the heart of California’s Death Valley, summer temperatures soar past 120°F (49°C). But while most life quickly collapses, one native plant is just okay with it impossible. Tidestromia oblongifolia, commonly called Arizona…
Continue Reading
-

Fireball meteor captured streaking through Welsh sky
The rare phenomenon was spotted from a residential home
An amateur astronomer was left astonished after witnessing a rare fireball…
Continue Reading
-

This is the largest, most detailed radio image yet of our Milky Way
array: A broad and organized group of objects. Sometimes they are instruments placed in a systematic fashion to collect information in a coordinated way. Other times, an array can refer to things that are laid out or displayed in a way that…
Continue Reading
-
Massive underwater volcano expected to erupt this year as risk to life explained – LADbible
- Massive underwater volcano expected to erupt this year as risk to life explained LADbible
- A Giant Volcano Off The Coast Of Oregon Is Scheduled To Erupt In 2026, JWST Finds The Best Evidence Yet Of A Lava World With A Thick Atmosphere, And Much…
Continue Reading
