Sinnatamby, C. S. Last’s Anatomy: Regional and Applied 12th edn (Churchill Livingstone, 2011).
Rogers, A. W. Textbook of AnatomyChurchill Livingstone,. (1992).
Palastanga, N. & Soames, R. Anatomy and Human Movement: Structure and Function 6th edn…
Sinnatamby, C. S. Last’s Anatomy: Regional and Applied 12th edn (Churchill Livingstone, 2011).
Rogers, A. W. Textbook of AnatomyChurchill Livingstone,. (1992).
Palastanga, N. & Soames, R. Anatomy and Human Movement: Structure and Function 6th edn…
Hou, Y., Wang, H., Fu, Y., Gao, K. & Zhang, H. Multi-objective brain storm optimization for integrated scheduling of distributed flow shop and distribution with maximal processing quality and minimal total weighted earliness and tardiness….

“What can you do with 1,000 neurons?” That’s the challenge driving a competition launched in July by computational neuroscientist Nicolas Rougier. Competitors score points by designing model brains to solve a series of simple…
In the rapidly advancing field of cellular biology, mitophagy has emerged as a pivotal process influencing both health and disease. A groundbreaking study recently published in Cell Research brings to light profound insights into the role…

Rising seas and saltier soils have placed Florida’s cabbage palm at great risk.
Even plants long considered resilient may struggle to survive as pollution continues to raise global temperatures.
A study…
BEIJING — Chinese scientists have successfully achieved the first direct mass measurement of a free-floating object, confirming that it is a planet with a mass comparable to that of Saturn.
This key research finding,…
BEIJING, Jan. 4 (Xinhua) — Chinese scientists have successfully achieved the first direct mass measurement of a free-floating object, confirming that it is a planet with a mass comparable to that…
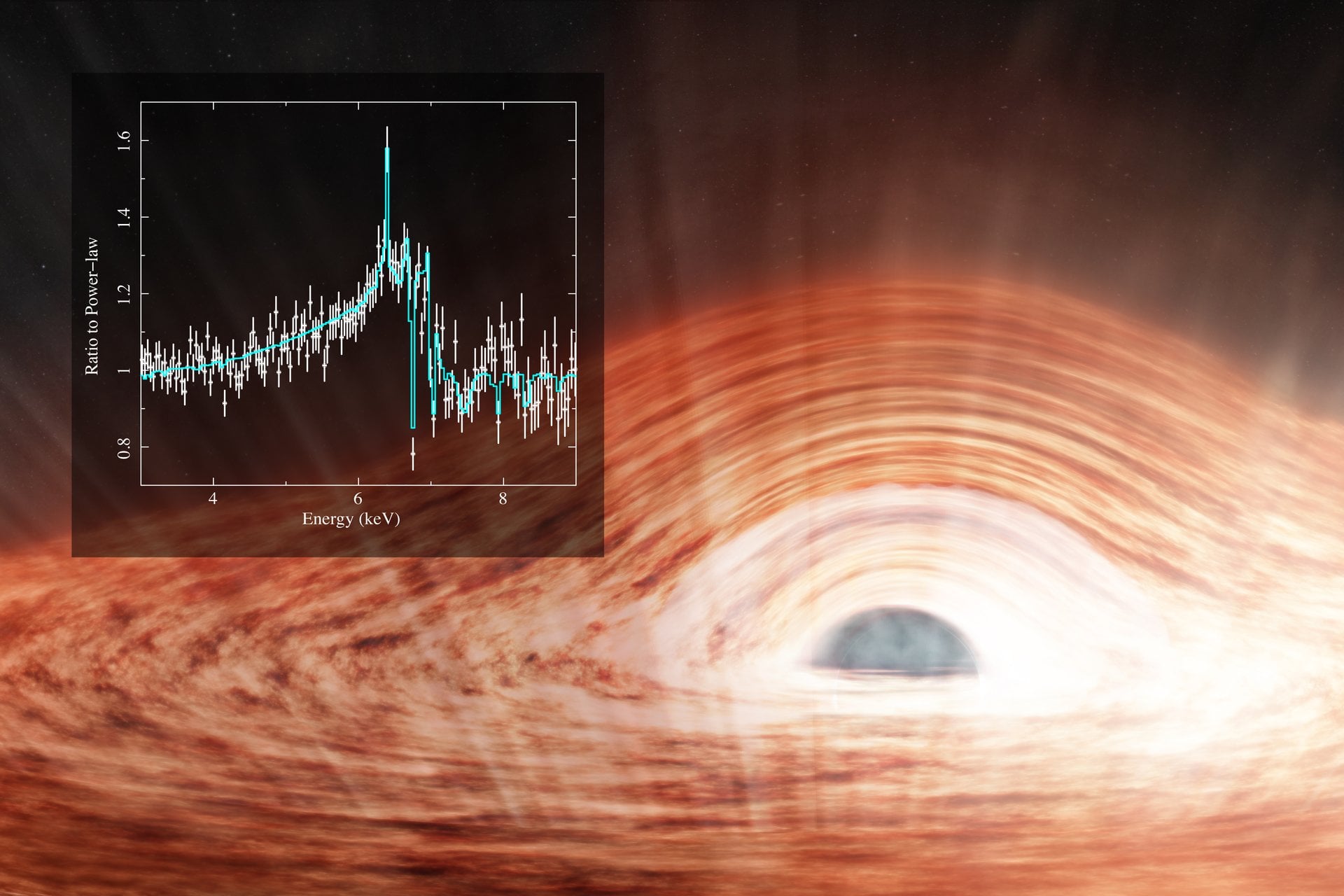
The X-Ray Imaging and Spectroscopy Mission (XRISM), a joint mission between the Japanese Aerospace Exploration Agency (JAXA) and NASA, launched on Sept. 7th, 2023. Its advanced imaging filters and spectrometers were designed to study…