On a frigid orbit beyond Neptune, some of the solar system’s smallest worlds project a strange silhouette. Two rounded lobes, pressed together with a narrow “neck,” like a snowman that never melted.
Those shapes are common enough to demand…
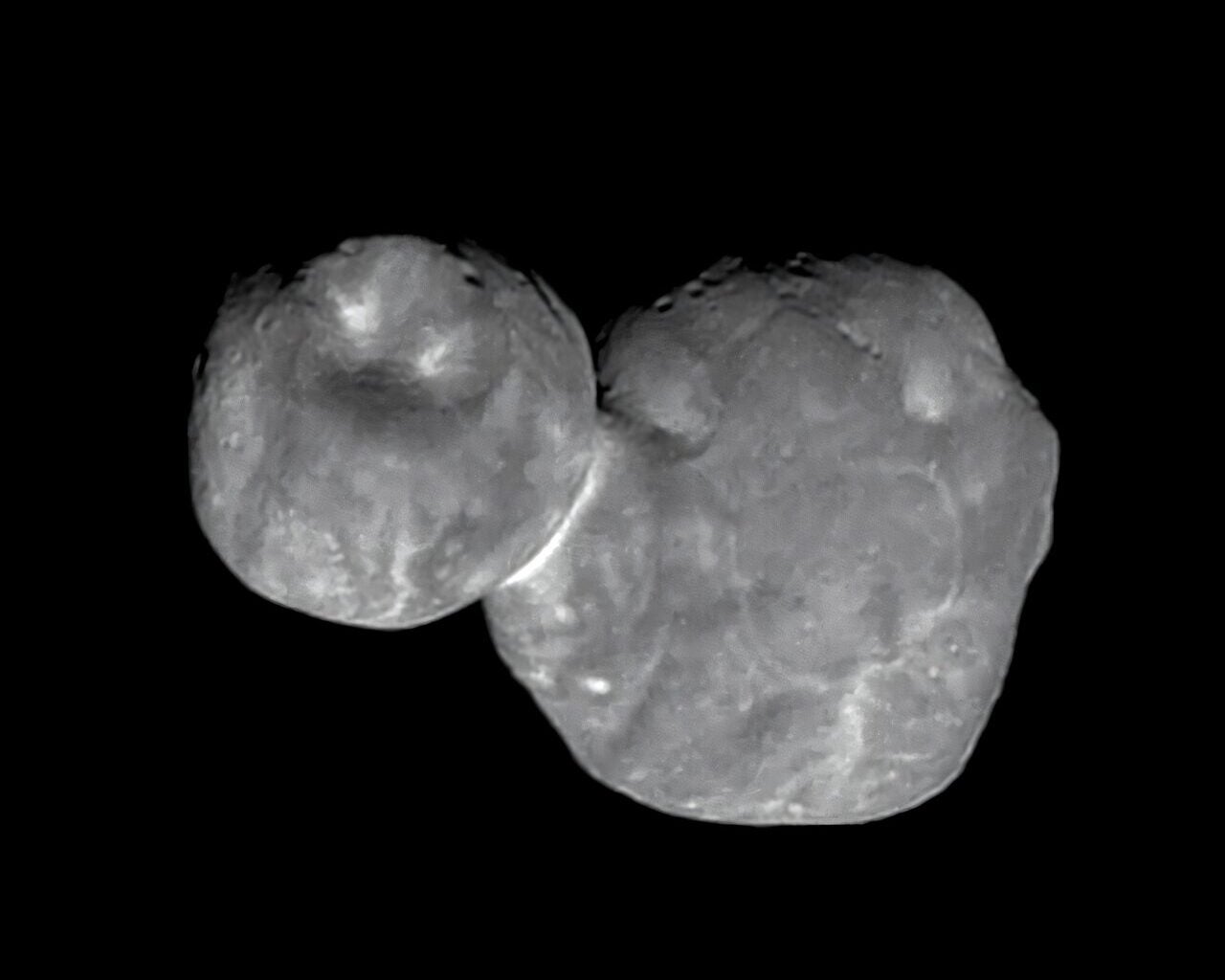
On a frigid orbit beyond Neptune, some of the solar system’s smallest worlds project a strange silhouette. Two rounded lobes, pressed together with a narrow “neck,” like a snowman that never melted.
Those shapes are common enough to demand…

When you step outside on a winter morning or pop a mint into your mouth, a tiny molecular sensor in your body springs into action, alerting your brain to the sensation of cold. Scientists have now captured the first detailed images…

It looks like a March launch is no longer in the cards for Artemis II, NASA’s first crewed trip to the moon’s vicinity since the final Apollo mission over 50 years ago. While preparations were underway at the Kennedy Space Center for a launch as…

Scientists have found traces of a tough material that once formed the outer shells of ancient sea creatures called trilobites, preserved inside fossils that are more than 500 million years old.
The discovery shows that parts of living organisms…
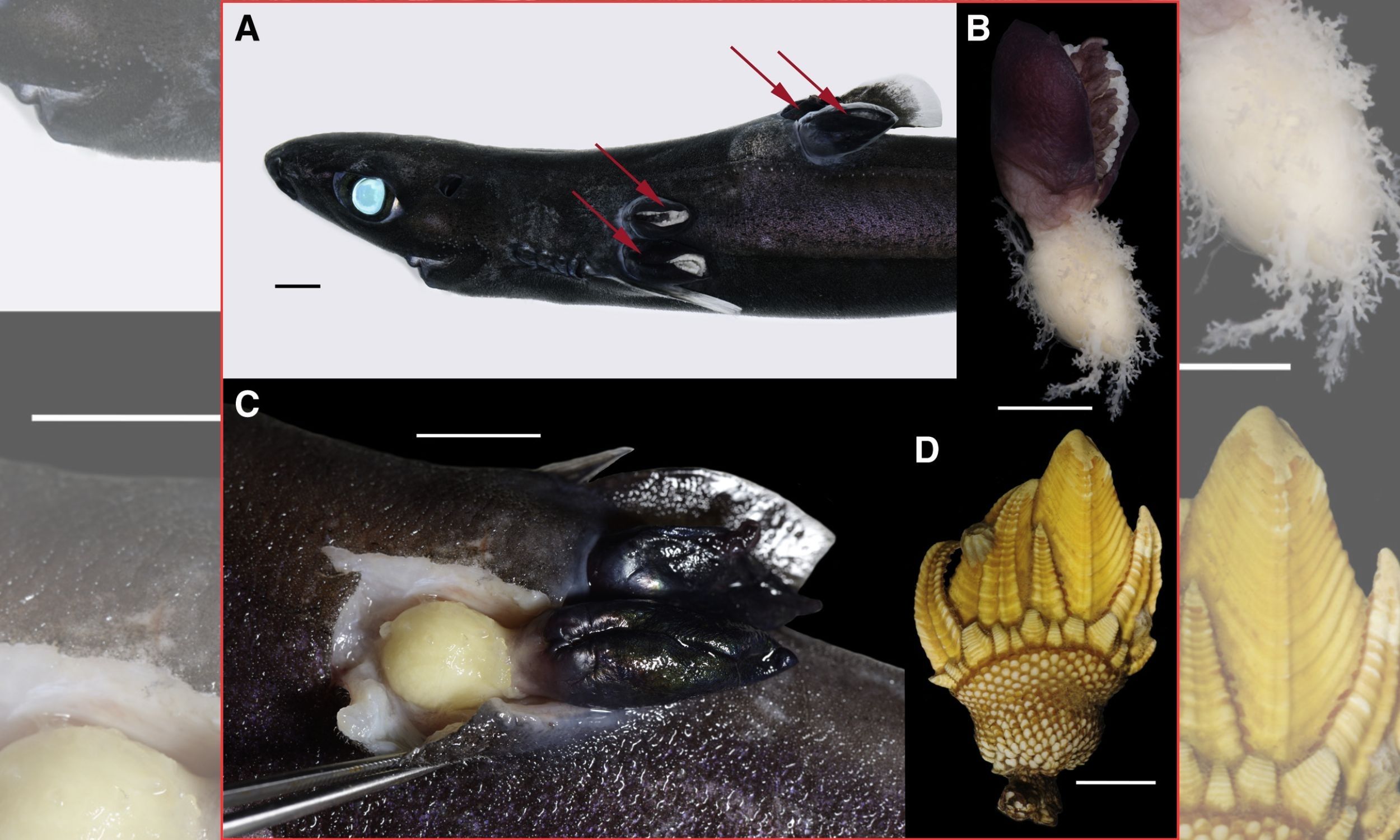
A common shoreline barnacle has been documented piercing deep-sea sharks and extracting nutrients directly from their flesh, marking a complete transition from filter feeding to parasitism.
That shift captures an evolutionary turning point in…