Six planets, Mercury, Venus, Saturn, Jupiter, Uranus, and Neptune will be visible along a sweeping curve in the sky tonight. While alignments occur periodically — the last six-planet one was in January 2025, followed by a four-planet display in…
Category: 7. Science
-

Six planets will be visible in a planetary parade tonight
Six planets, Mercury, Venus, Saturn, Jupiter, Uranus, and Neptune will be visible along a sweeping curve in the sky tonight. While alignments occur periodically — the last six-planet one was in January 2025, followed by a four-planet display in…
Continue Reading
-
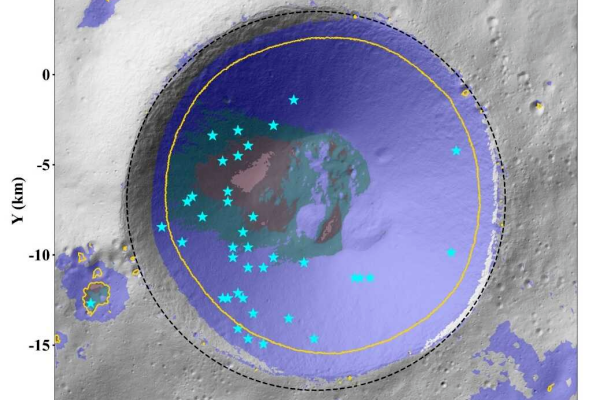
Chinese scientists map ice ‘treasure map’ for Chang’e 7 mission at moon’s south pole
BEIJING — Chinese scientists have developed a high-resolution model to assess the thermal stability of water ice in the Shackleton Crater region near the lunar south pole, providing crucial guidance for the Chang’e 7…
Continue Reading
-

Chinese scientists map ice ‘treasure map’ for Chang’e 7 mission at moon’s south pole
BEIJING — Chinese scientists have developed a high-resolution model to assess the thermal stability of water ice in the Shackleton Crater region near the lunar south pole, providing crucial guidance for the Chang”e 7…
Continue Reading
-

A total lunar eclipse will turn the moon blood red on Tuesday across several continents – Arab News PK
- A total lunar eclipse will turn the moon blood red on Tuesday across several continents Arab News PK
- Where to see the total lunar eclipse in the early hours of March 3 Space
- The Eerie ‘Blood’ Moon Will Grace the Night Sky Next Week, Thanks to…
Continue Reading
-
A total lunar eclipse will turn the moon blood red on Tuesday across several continents – Arab News
- A total lunar eclipse will turn the moon blood red on Tuesday across several continents Arab News
- Where to see the total lunar eclipse in the early hours of March 3 Space
- The Eerie ‘Blood’ Moon Will Grace the Night Sky Next Week, Thanks to a…
Continue Reading
-
Total lunar eclipse brings blood moon on March 3 – WLIW
- Total lunar eclipse brings blood moon on March 3 WLIW
- The 3 March 2026 Total Lunar Eclipse: a live, international event The Virtual Telescope Project 2.0
- Want to see the total lunar eclipse? Here’s the gear you’ll need for March. Live Science
Continue Reading
-

Nasa announces Artemis III mission no longer aims to send humans to moon | Nasa
Nasa announced on Friday radical changes to its delayed Artemis III mission to land humans back on the moon, as the US space agency grapples with technical glitches and criticism that it is trying to do too much too soon.
The abrupt shift in…
Continue Reading
-

A total lunar eclipse will turn the moon blood red on Tuesday across several continents
A blood-red moon will soon grace the skies for a total lunar eclipse — and there won’t be another until late 2028.
The spectacle will be visible Tuesday morning from North America, Central America and the western part of South America….
Continue Reading
-

Six planets due to parade across night sky in rare celestial spectacle | Astronomy
Six planets will parade across the sky this weekend in a rare celestial spectacle, experts have said.
For the next few days, Jupiter, Saturn, Venus, Mercury, Neptune and Uranus will all be visible at the same time in the night sky – although…
Continue Reading