Human language is remarkably rich and intricate. Yet from the standpoint of information theory, the same ideas could theoretically be transmitted in a far more compressed format. That raises an intriguing question: why do people not communicate…
Category: 7. Science
-
DNA Reveals Identity of Britain’s 11,000-Year-Old “Oldest Northerner” – SciTechDaily
- DNA Reveals Identity of Britain’s 11,000-Year-Old “Oldest Northerner” SciTechDaily
- News – Remains of England’s Earliest Known “Northerner” Belong to Mesolithic Girl Archaeology Magazine
- Northern Britain’s oldest human remains are of a young…
Continue Reading
-

Australia launches research to develop TR4-resistant banana varieties
A new research project aims to help protect banana crops from Fusarium wilt Tropical Race 4 (TR4), a soil-borne disease affecting global banana production.
The project is funded by Hort Innovation and will use genetic tools to…
Continue Reading
-
Chinese scientists develop AI model to push deep-space exploration-Xinhua
BEIJING, Feb. 20 (Xinhua) — Chinese researchers have developed an artificial intelligence (AI) model for astronomical imaging that significantly enhances scientists’ ability to peer into the deepest reaches of the cosmos.
A…
Continue Reading
-

Gaia Detected an Entire Swarm of Black Holes Traveling Through The Milky Way : ScienceAlert
A fluffy cluster of stars spilling across the sky may have a secret hidden in its heart: a swarm of over 100 stellar-mass black holes.
The star cluster in question is called Palomar 5. It’s a stellar stream that stretches out across 30,000…
Continue Reading
-

Fossils of new Spinosaurus species found in the Sahara
At a remote and barren Sahara desert site in Niger, scientists have unearthed fossils of a new species of Spinosaurus, among the biggest of the meat-eating dinosaurs, notable for its large blade-shaped head crest and jaws bearing interlocking…
Continue Reading
-
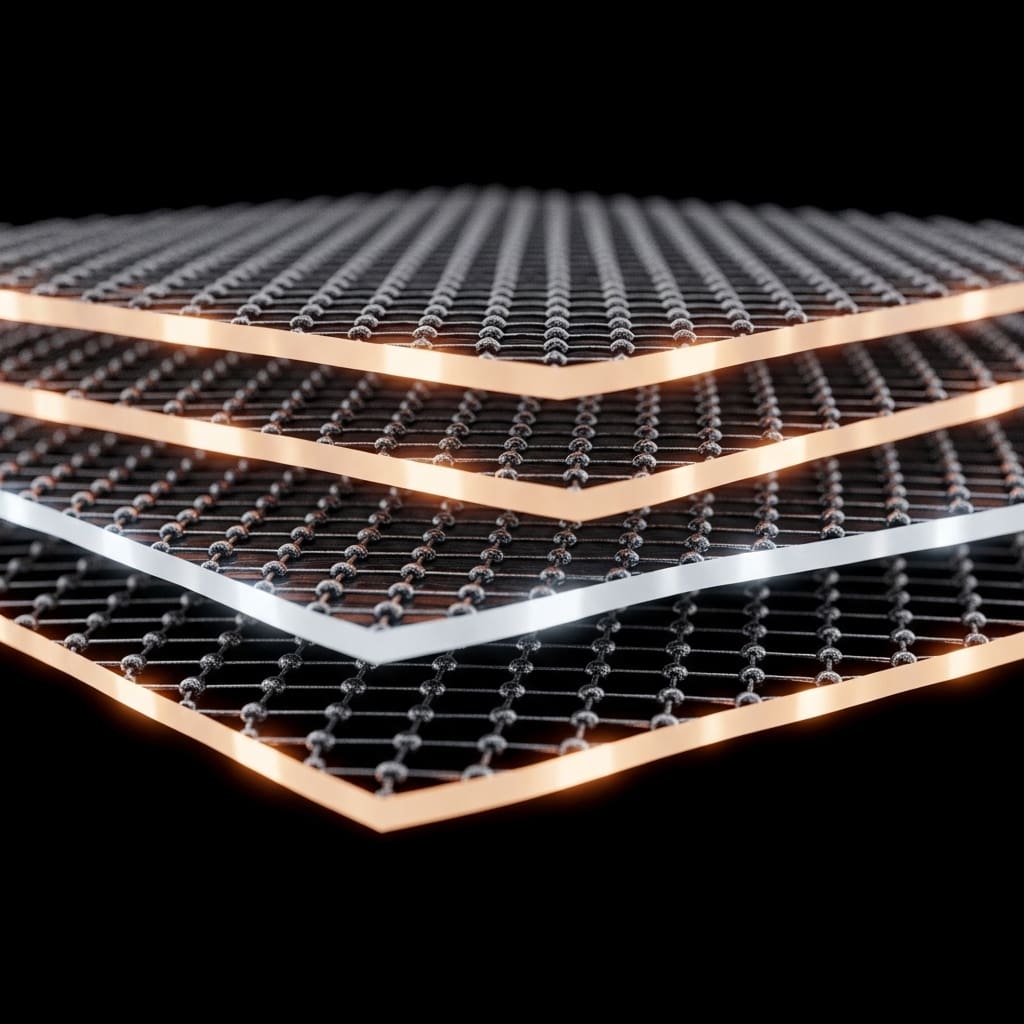
Graphene Patterns Unlock Stacked Insulating Electron States
Researchers are increasingly focused on harnessing correlated insulating behaviour in graphene to unlock emergent phenomena such as superconductivity and magnetism. Xinyu Cai, Fengfan Ren, and Qiao Li, all from ShanghaiTech University,…
Continue Reading
-
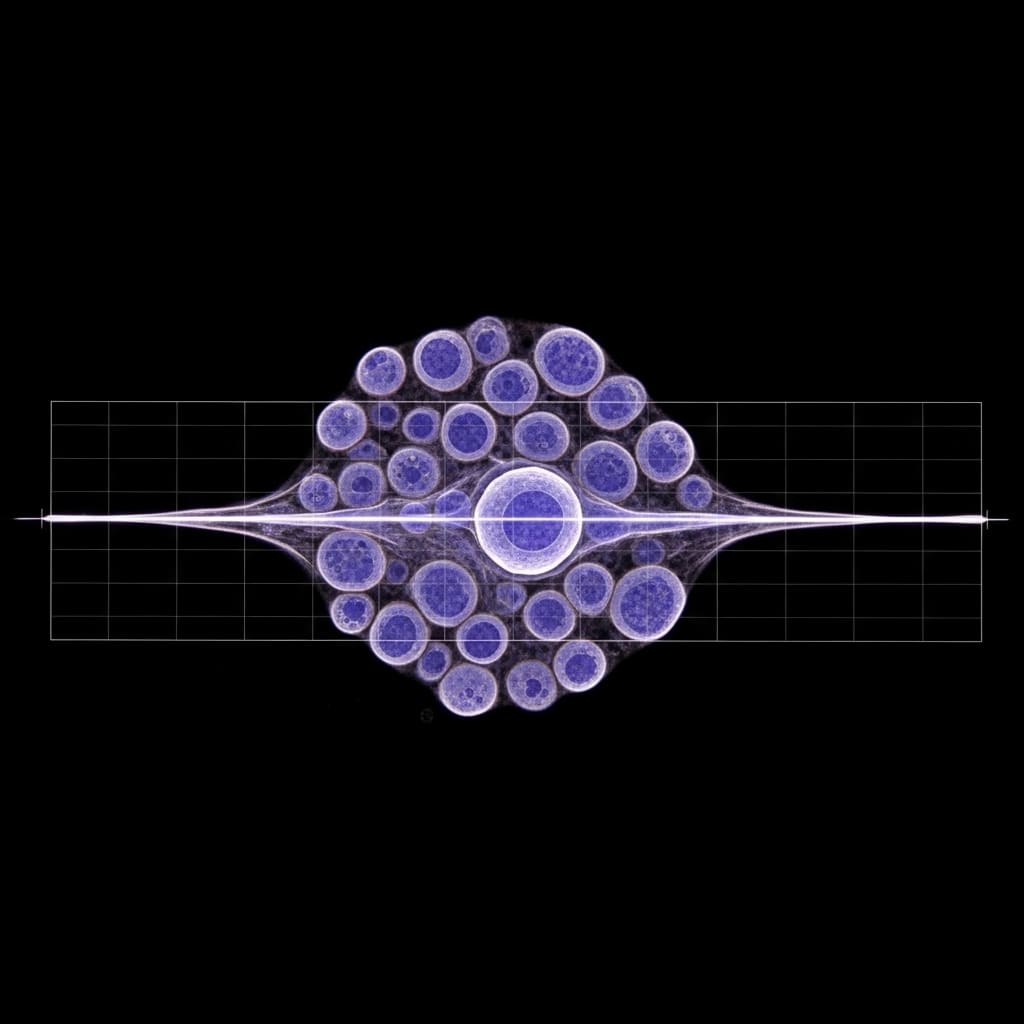
Simple Methods Match Complex AI For Cell Analysis
Single-cell RNA sequencing data consistently reveal robust statistical structures, prompting the creation of complex foundation models like TranscriptFormer to generate gene expression embeddings. Huan Souza and Pankaj Mehta, both from the…
Continue Reading
-

Literary Hub » What Growing Up On Mars Would Do to the Human Body
The annual SXSW conference in Austin, Texas, can be a bit overwhelming. What started as a fairly modest four-day music festival in 1987, drawing some 700 attendees, has become a ten-day extravaganza of panel presentations featuring…
Continue Reading
-
Chinese researchers propose new explanation for ultra-long gamma-ray burst-Xinhua
BEIJING, Feb. 20 (Xinhua) — Chinese researchers have developed a novel model to explain the origin of an ultra-long gamma-ray burst (GRB), challenging the conventional understanding of these violent cosmic events.
Gamma-ray bursts are among…
Continue Reading
