A “house of cards” is such a wonderful English phrase, one now primarily associated with a Netflix political drama. However, its original meaning refers to a fundamentally unstable system.
It’s also the term Sarah Thiele, originally a PhD…

A “house of cards” is such a wonderful English phrase, one now primarily associated with a Netflix political drama. However, its original meaning refers to a fundamentally unstable system.
It’s also the term Sarah Thiele, originally a PhD…

Watch On
Europe’s towering Ariane 6 rocket is gaining momentum in the heavy-lift launch market as the vehicle gears up for its fifth flight.

Earlier this year, leaked budget cuts cast a dark shadow over the future of NASA’s Nancy Grace Roman Space Telescope—a multi-billion-dollar instrument with the capacity of “200 Hubbles,” according to experts. Thankfully,…

In 2024, the reef experienced its most widespread coral-bleaching event on record, affecting the entire reef system. By 2025, surveys by the Australian Institute of Marine Science (AIMS) documented the largest annual drop in live coral…

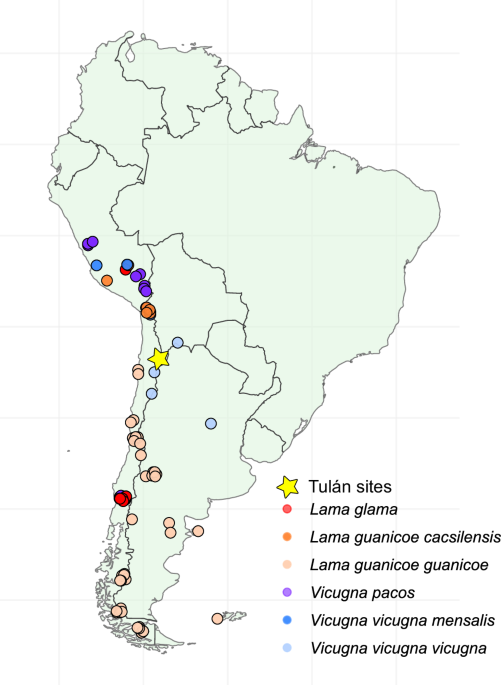
All research was conducted in accordance with Chilean regulations. Excavation permits were issued by the Consejo de Monumentos Nacionales de Chile (CMN) under permit N° 4409 (18.11.2013). Export of osteological samples…
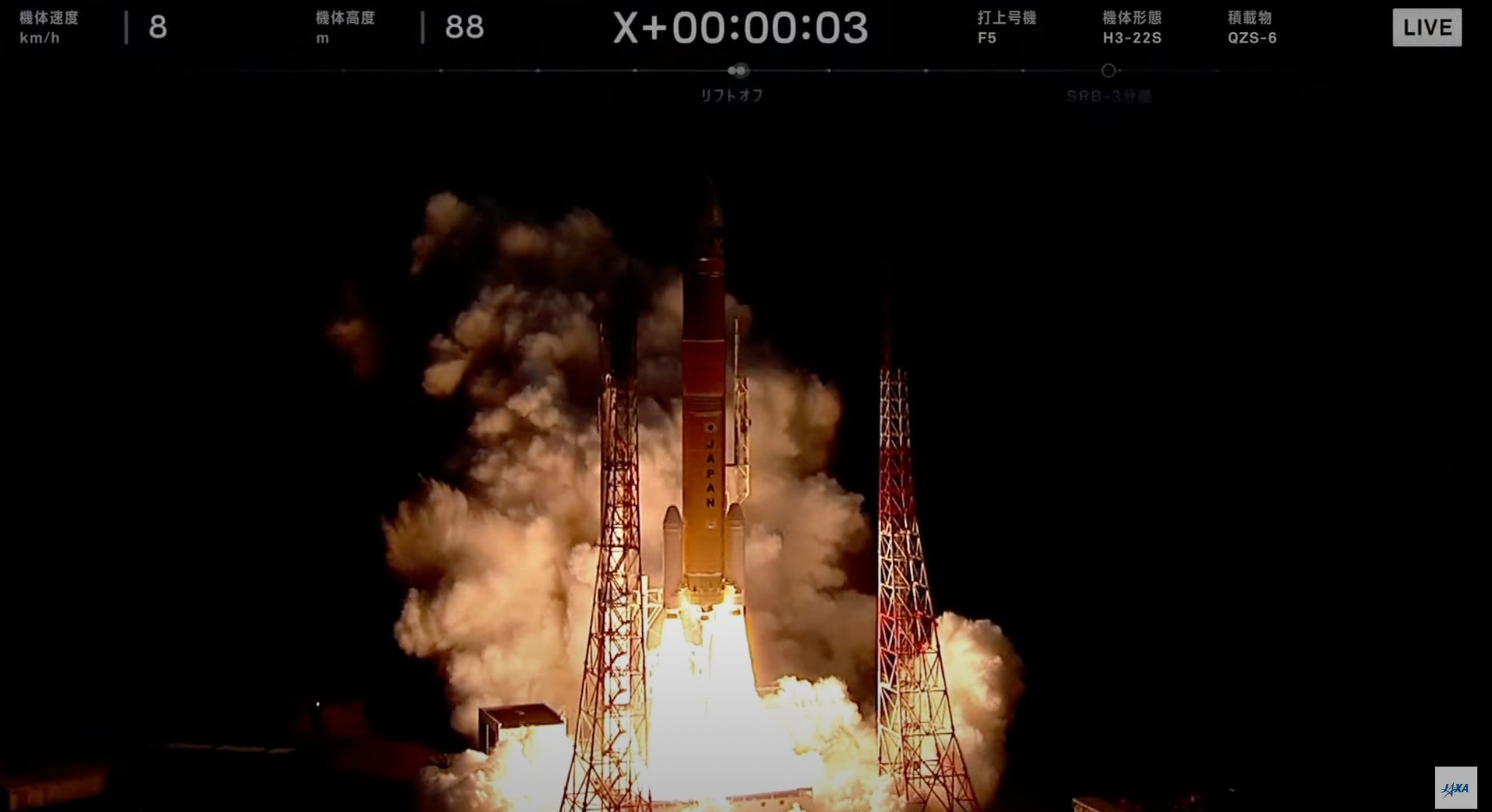
Watch On
Japan will launch a new navigation satellite to orbit tonight (Dec. 16), and you can watch the action live.
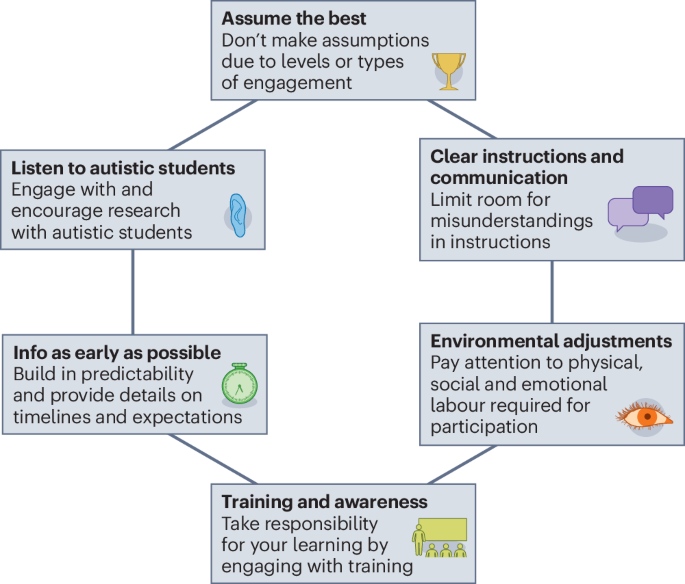
Autism is a lifelong condition impacting how individuals process information and interact with the world, simultaneously protected as a disability and recognized as a form of neurodivergence. Autism affects how people think, feel and communicate…

Dungee, R., Greene, O., Schonhut-Stasik, J. et al. Neurodivergent in astronomy: the early-career researcher edition.
Nat Astron 9, 1754–1757 (2025). https://doi.org/10.1038/s41550-025-02742-0