Every biologist knows how important the Great Oxygenation Event was. It took the first photosynthetic organisms hundreds of millions of years to enrich Earth’s atmosphere with oxygen, leading to complex life like us. But before complex,…
Category: 7. Science
-
ELDGG: an end-to-end LiDAR-dynamic-guided GAN for hyperspectral image hierarchical reconstruction and classification
Gao, H. et al. AMSSE-Net: Adaptive multiscale spatial–spectral enhancement network for classification of hyperspectral and LiDAR data. IEEE Trans. Geosci. Remote Sens. 61, 1–17 (2023).
Li,…
Continue Reading
-
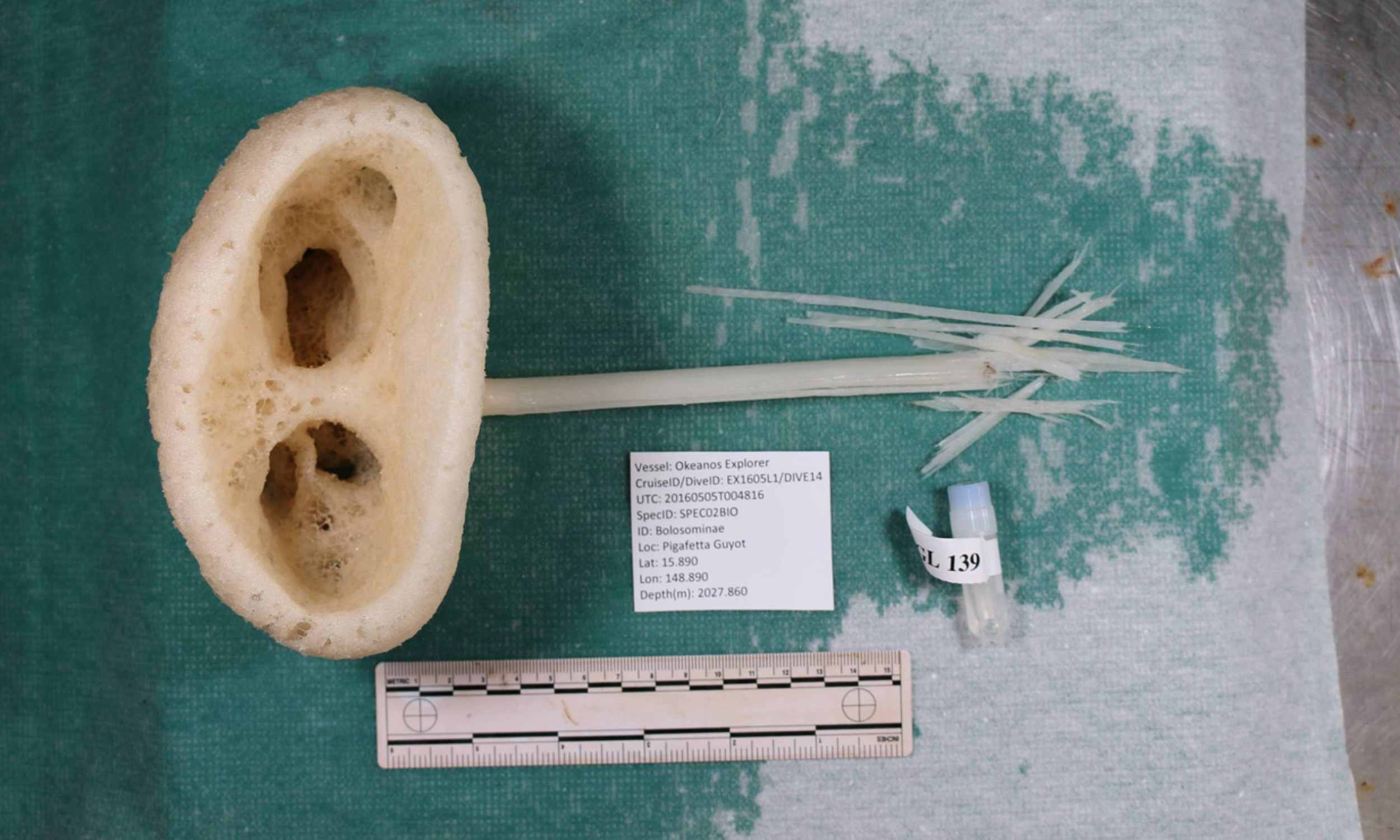
Strange new sponge species is given the name “magnificent alien”
Far below the Pacific waves, nearly 6,560 feet down, a pale sponge on a thin stalk lives on a cold, dark slope. This new sponge was given the scientific name Advhena magnifica, which means “magnificant alien,” which is an apt…
Continue Reading
-

Multi-University Hypersonics Research Initiative Launches at UCF
Leading researchers from across the country gathered at UCF on Dec. 12 to launch a major Multi University Research Initiative (MURI), supported by the U.S. Army Research Office, aimed at transforming how scientists understand and…
Continue Reading
-

Amendment 30: Updates to the ROSES-25 Summary of Solicitation
ROSES-2025 is an omnibus or umbrella solicitation that contains many program element appendices (listed in Tables 2 and 3) and the ROSES-25 Summary of Solicitation (SoS) lays out the backstop rules that apply by default to those program…
Continue Reading
-
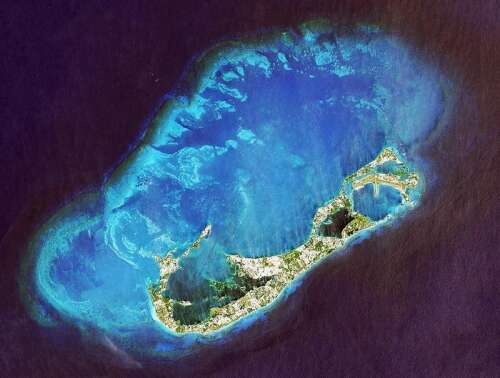
Giant rock mass beneath island hints at ancient origins – The Royal Gazette
Created: Dec 15, 2025 03:16 PM
What lies beneath: a satellite view of Bermuda and the surrounding reef platform. The island sits on a vast intrusion of rock pushing up the oceanic crust (File photograph)
Scientific…
Continue Reading
-

What babies’ brains teach us about development
A lot of brain development happens early in life, but researchers don’t have a strong understanding of how a baby’s brain develops while they’re awake.
New research from Northeastern University sheds light on how babies develop…
Continue Reading
-

Interstellar comet 3I/Atlas will make closest approach to Earth on Friday
CAPE CANAVERAL, Fla. (AP) — A stray comet from another star swings past Earth this week in one last hurrah before racing back toward interstellar space.
Discovered over the summer, the comet known as 3I/Atlas will pass…
Continue Reading
-

World heading toward ‘peak glacier extinction’ with up to 4,000 set to disappear a year
BERN – Hundreds gathered to say goodbye when 700-year-old Pizol died. The funeral in Switzerland in 2019 was solemn. Mourners wore black; flowers were laid; a priest spoke. It was a symbolic moment: Pizol had been a glacier, but human-driven…
Continue Reading
-

Beyond the ground station: Why space data centers require a distributed RAID
Low Earth orbit is undergoing the fastest expansion in its history. However, the nature of the payload is changing. We are moving past simple “bent-pipe” communication relays — where a satellite simply receives a signal and beams it…
Continue Reading
