Scientists are increasingly focused on resolving the singularities predicted by classical general relativity through the framework of loop quantum gravity. Xiaotian Fei and Cong Zhang, both from the School of Physics and Astronomy, Key…
Category: 7. Science
-
NASA Puts Starliner Mishap in Same Class as Shuttle Tragedies – Bloomberg.com
- NASA Puts Starliner Mishap in Same Class as Shuttle Tragedies Bloomberg.com
- NASA Thinks Boeing’s Starliner Can Fly in April, Which Is Hilarious Gizmodo
- “Great Progress”: Boeing Stock (NYSE:BA) Plunges as Starliner Remains a Work in…
Continue Reading
-
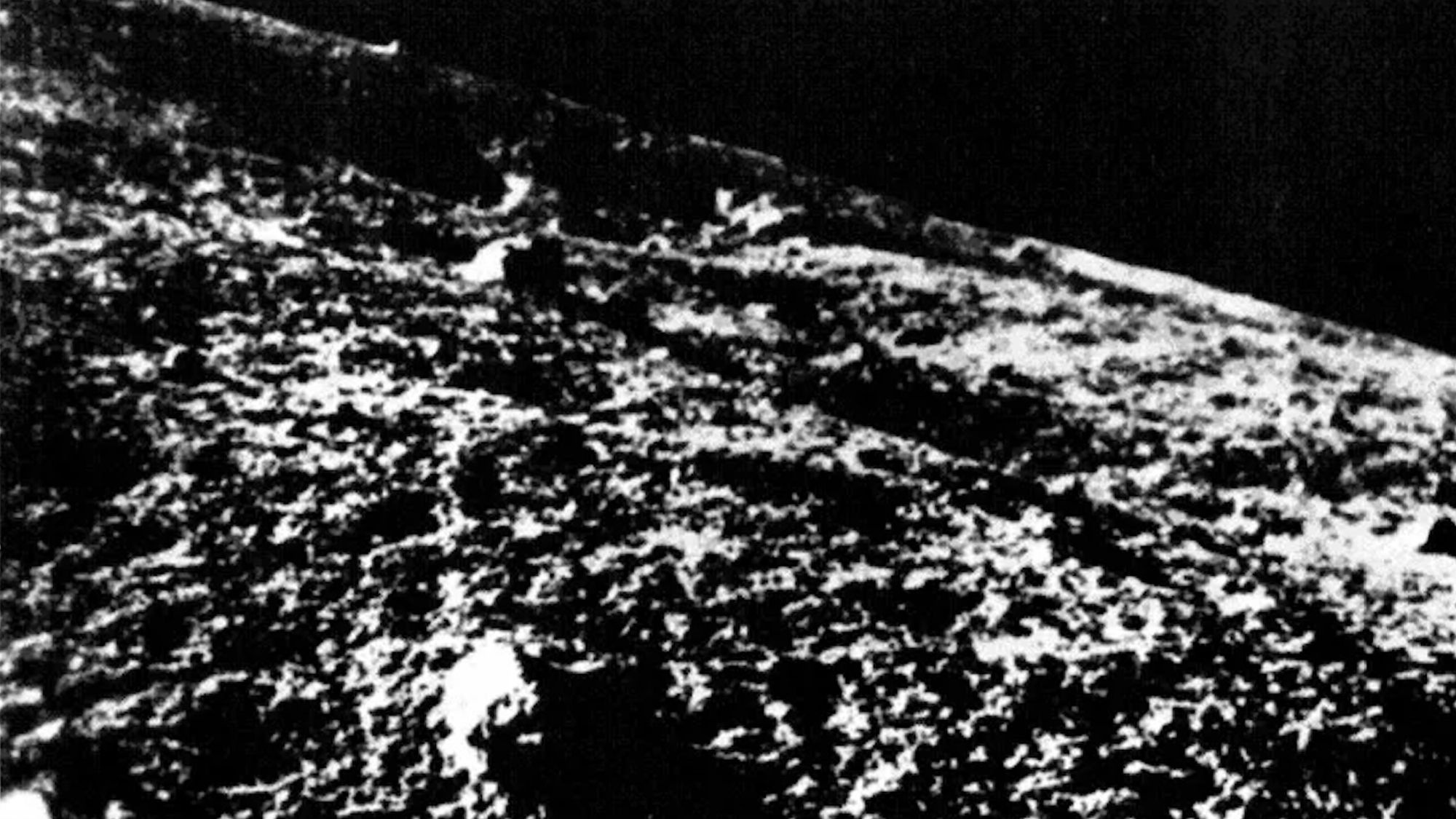
AI helps find new clues in the 60-year search for Luna 9, the 1st successful moon lander
Scientists may be closing in on the long-missing landing site of the Soviet Union’s Luna 9, the first human-made object to safely touch down on the moon.
On Feb. 3, 1966, Luna 9 descended into Oceanus Procellarum on the western edge of the…
Continue Reading
-

Map the Earth’s Magnetic Shield with the Space Umbrella Project
A stream of charged particles known as the solar wind flows from the Sun toward Earth. Here, it meets the Earth’s magnetic fields, which shield our planet like a giant umbrella. The Space Umbrella project needs your help investigating this…
Continue Reading
-

Warming Arctic leaves ringed seals vulnerable to toxic buildup
A single unusually warm year can significantly affect the health of Arctic animals. A new study shows that Arctic ringed seals are facing a double threat: climate-driven nutritional stress and a buildup of harmful contaminants in their bodies….
Continue Reading
-
NASA conducts second rocket fueling test that will decide when Artemis astronauts head to the moon – The Washington Post
- NASA conducts second rocket fueling test that will decide when Artemis astronauts head to the moon The Washington Post
- Artemis II Wet Dress Rehearsal: Countdown Begins NASA (.gov)
- NASA to rehearse moon launch again after repairing its rocket NBC…
Continue Reading
-
Inhibitory effect of the methanol extract of Clerodendrum volubile Linn. on carbon tetrachloride-induced early hepatic fibrogenesis: in silico and in vivo study
Devarbhavi, H. et al. Global burden of liver disease: 2023 update. J. Hepatol. 79, 516–537 (2023).
Friedman, S. L. Molecular regulation of hepatic fibrosis, an integrated cellular response to…
Continue Reading
-
The History of Proteomics: Key Milestones and Innovations
Proteomics—the large-scale study of the entire set of proteins produced by a biological system—has evolved over several decades. Its development was shaped by advances in separation technologies, mass spectrometry, and bioinformatics….
Continue Reading
-

Turtle brains reveal ancient visual processing
For decades, scientists assumed that sophisticated visual abilities were largely unique to mammals. They believed that only large, folded cerebral cortices could reliably detect and interpret important changes in the environment.
However, new…
Continue Reading
-

The investigation could solve the mystery of how supermassive black holes grew so large in the early universe.
Using NASA’s Hubble Space Telescope and Chandra X-ray Observatory, astronomers have hunted for “wandering” black holes drifting through dwarf galaxies. The discovery of these rogue black holes in such small galaxies could provide a “fossil…
Continue Reading
