Certain neighborhood characteristics, including higher poverty, more uninsured residents, and lower educational attainment, may lead to an increase in COPD-related emergency department visits and hospitalizations, according to a new…
Category: 7. Science
-
Do you suffer from IBS? This doctor says ‘gravity intolerance’ may be to blame
Gravity, one of the four fundamental forces of nature, keeps us down to Earth (literally), but our body’s relationship to it could explain our susceptibility to some common health conditions — for instance, irritable bowel syndrome.
At least,…
Continue Reading
-

Quantum ‘magic’ Linked To Critical Shifts In Systems
Scientists are increasingly recognising nonstabilizerness, a key resource for universal computation, as crucial to understanding complex quantum systems. Andrew Hallam, Ryan Smith, and Zlatko Papić, all from the School of Physics and…
Continue Reading
-

Scientists discover mouse gene that could determine if dads are doting or angry
A specific brain region, dubbed a “parenting hub”, has been linked to the diverse paternal…
Continue Reading
-

Sensors Information | AZoSensors.com – Page not found
Terms
While we only use edited and approved content for Azthena
answers, it may on occasions provide incorrect responses.
Please confirm any data provided with the related suppliers or
…Continue Reading
-
The Sky Today on Thursday, February 19: Mercury at greatest eastern elongation – Astronomy Magazine
- The Sky Today on Thursday, February 19: Mercury at greatest eastern elongation Astronomy Magazine
- Crescent Moon, Venus and Mercury The Vineyard Gazette
- Rare Chance to See Mercury: Planet Will Be Visible in the Evening Sky for Only Two Days
Continue Reading
-
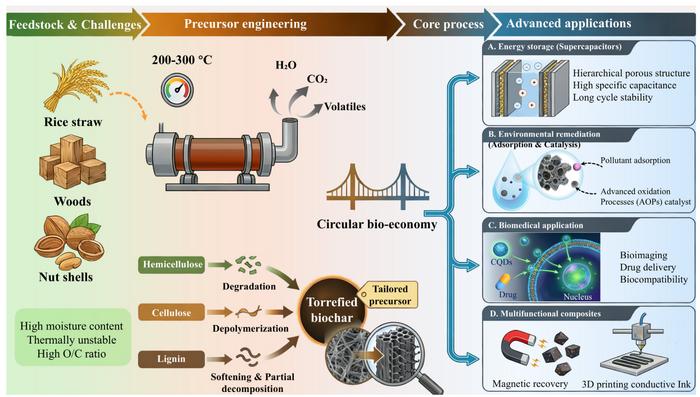
The process powering future carbon materials
A comprehensive review of biomass torrefaction as a versatile platform for the synthesis of functional carbon materials image: ©
Wei Han, Yifan Wang, Lei Wang, Peng Xie, Tianqi Liu, Qinglian Wu, Chunshuang Zhou, Xiaomeng Guo, Lina…Continue Reading
-

Brain development may continue into your 30s, new research shows
Scroll through TikTok or Instagram and you will likely see the familiar line: “Your frontal lobe isn’t fully developed yet.” It has become a popular explanation for questionable choices, from ordering another drink to texting someone you…
Continue Reading
-
Researchers capture 1st evidence of Australian sea lion mom-pup foraging lessons-Xinhua
CANBERRA, Feb. 19 (Xinhua) — Researchers have found that Australian sea lion pups learn how to hunt from their mothers, marking the first evidence of social transmission of foraging behavior in these marine mammals.
Social information…
Continue Reading
-

Turkish, Bulgarian scientists uncover Antarctica’s geological history spanning millennia
HORSESHOE ISLAND, Feb 19 (BelTA/APP): Turkish and Bulgarian geologists participating in Türkiye’s 10th National Antarctic Science Expedition have launched a joint research initiative to uncover Antarctica’s geological history and assess its…
Continue Reading