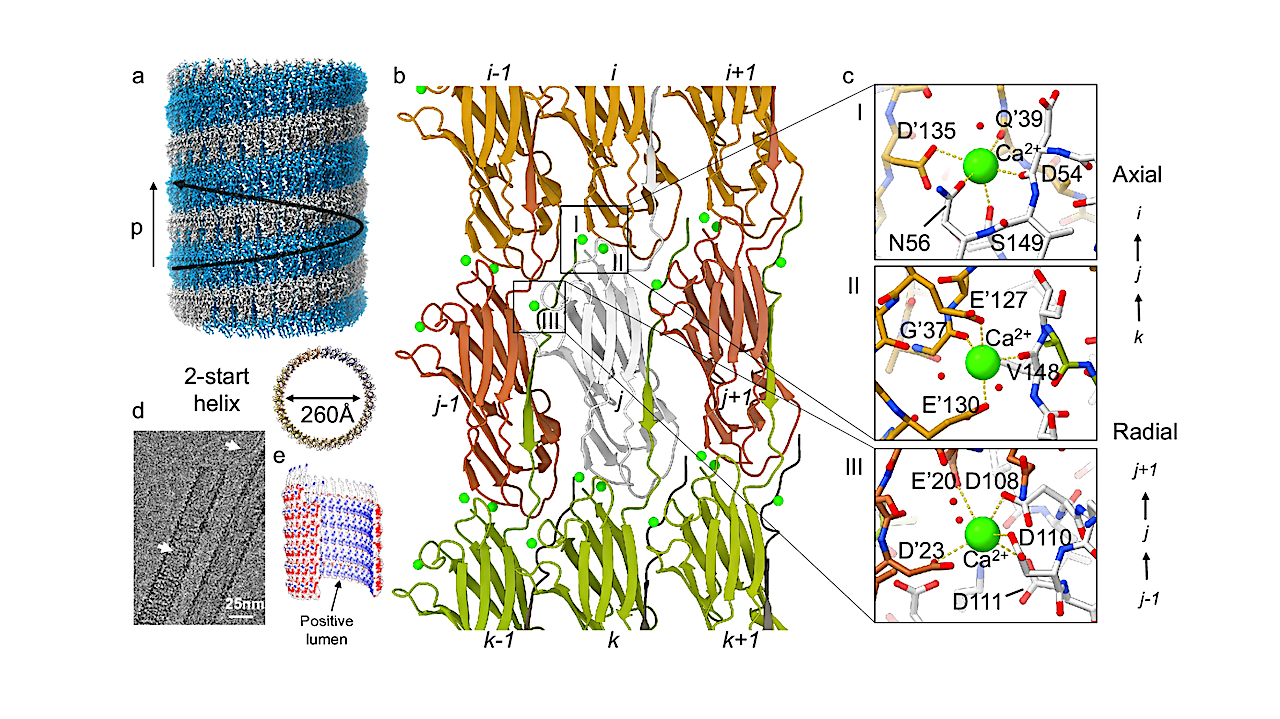
On a routine wildlife survey in a forest reserve in southwest China, scientists caught a weasel that turned out to be a species no one had recorded before. The animal, now named Mustela mopbie, is the newest member of a famously elusive group of…
Category: 7. Science
-

Scientists make breakthrough that could give major boost to food supply: ‘A huge positive’
Scientists at the University of Guelph in Ontario, Canada, have developed a gene editing technique that improves canola yield for farmers by introducing a particular starch-producing enzyme into the crop’s genetic material.
According to the…
Continue Reading
-

Killer sea sponge discovered that traps and devours live animals
Far beneath Antarctic waters, scientists have found a killer sea sponge with a near perfect ball shape that grabs and eats passing animals. It is the star of dozens of newly confirmed deep sea species from a remote part of the Southern Ocean.
A…
Continue Reading
-

New fish species discovered has an organ that is new to science
Scientists in the Colombian Amazon have described a new fish species called Priocharax rex, only about 0.8 inches long, in a 2025 study.
It is the largest known member of its tiny genus and carries a round wing-like flap of skin between its…
Continue Reading
-
First Results From The Subaru Telescope's OASIS Survey: Direct Imaging Of New Worlds Around Unexplored Stars – astrobiology.com
- First Results From The Subaru Telescope’s OASIS Survey: Direct Imaging Of New Worlds Around Unexplored Stars astrobiology.com
- Researchers discover new worlds around uncharted stars, provide critical target for NASA’s next space telescope UT…
Continue Reading
-
On the coincidence between the close passage of HD7977 and the Pliocene-Pleistocene transition
Oort, J. The structure of the cloud of comets surrounding the solar system and a hypothesis concerning its origin. Bull. Astron. Institutes Neth. 11, 91–110 (1950).
Whipple, F. L. A comet…
Continue Reading
-
Aurora map: NOAA issues strong geomagnetic storm watch for this week in the U.S. – Big Rapids Pioneer
- Aurora map: NOAA issues strong geomagnetic storm watch for this week in the U.S. Big Rapids Pioneer
- How the solar storms that cause the Northern Lights can wreak havoc on Earth BBC
- Strong (G3) Geomagnetic Storm WATCH Valid for 09 Dec 2025
Continue Reading
-

Lost lineage of early humans in Africa rewrites our origin story
Using samples from caves and rock shelters along South Africa’s coasts, scientists have analyzed the DNA of ancient hunter-gatherers.
The records reveal a southern African lineage of Homo sapiens that sits at one of the deepest – and most…
Continue Reading