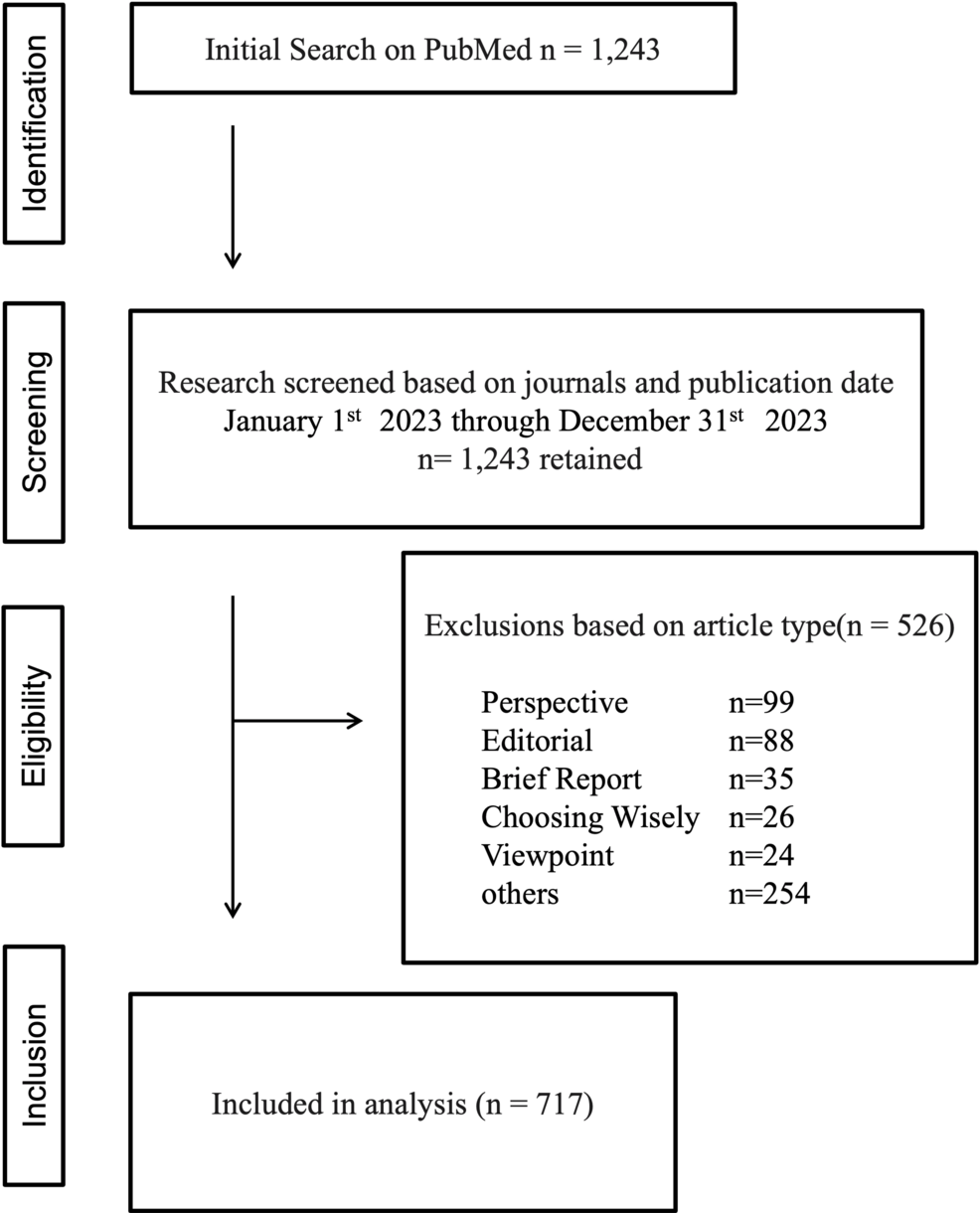
On Nov. 2, 2025, NASA honored 25 years of continuous human presence aboard the International Space Station. What began as a fragile framework of modules has evolved into a springboard for international cooperation, advanced scientific research…
Category: 7. Science
-

‘Cold Supermoon’ 2025: Why the final full moon of the year also towers highest
Skywatchers are in for a stunning spectacle this week when the second-biggest full moon of 2025, the Cold Supermoon, rises in the east at dusk and appears higher in the night sky than any other full moon of the year.
Officially full at 6:14 p.m….
Continue Reading
-
Coral reefs have helped stabilize Earth’s carbon cycle since ancient times: study-Xinhua
SYDNEY, Dec. 2 (Xinhua) — Coral reefs have played a vital role in conducting the rhythm of Earth’s carbon and climate cycles for more than 250 million years, a new study revealed.
Coral reefs have long been celebrated as biodiversity…
Continue Reading
-

JWST reveals ‘Alaknanda’ a massive spiral galaxy from the early universe
Image of the newly discovered spiral galaxy Alaknanda (inset) as observed in the shorter wavelength JWST bands. Several bright galaxies from the foreground Abell 2744 cluster are also seen. Credit
© NASA/ESA/CSA, I. Labbe/R….Continue Reading
-

New Quantum State May Propel Future Space Tech
Researchers at the University of California, Irvine have identified a previously unobserved form of quantum matter. According to the team, this state arises inside a specially engineered material that may one day support self-charging…
Continue Reading
-

New state of quantum matter could power future space tech
Researchers at the University of California, Irvine have identified a previously unobserved form of quantum matter. According to the team, this state arises inside a specially engineered material that may one day support self-charging computers…
Continue Reading
-

SpaceX completes record 60th orbital launch of 2025
SpaceX has set a new internal record by completing its 60th orbital launch mission of 2025, the highest number of flights the company has achieved in a single calendar year. A Falcon 9 rocket lifted off from Space Launch Complex 4E at Vandenberg…
Continue Reading
-

Moon-Jupiter duo, Geminid meteors, interstellar comet Earth visit
As the 2025 year draws to a close, the December night…
Continue Reading