National Aeronautics and Space Administration (NASA) launched two suborbital sounding rocket missions from Alaska this week to assess the…
ⓘ Jack B via Unsplash
The…

National Aeronautics and Space Administration (NASA) launched two suborbital sounding rocket missions from Alaska this week to assess the…

Jared Isaacman, an administrator at NASA, said in a post on X that astronauts will be allowed to bring their iPhones in space missions soon. In the past, astronauts were not permitted to bring their smart devices during flights, but some…

ⓘ Jack B via Unsplash
The…
Saturn’s largest moon may be the surviving remnant of a catastrophic collision between two earlier moons.
A new study reframes Titan not as a quiet survivor from the Solar System’s birth, but as the product of a comparatively recent collision…

For a few brief nights each year, you get a rare chance to watch a monster blink.
The Event Horizon Telescope collaboration has released new, detailed views of M87*, the supermassive black hole at the heart of the galaxy M87. The images do not…
Ocean plastic pollution is one of the greatest environmental challenges facing our planet today. The vastness of the oceans, combined with the complex nature of marine debris, makes it difficult to monitor and address effectively. However,…
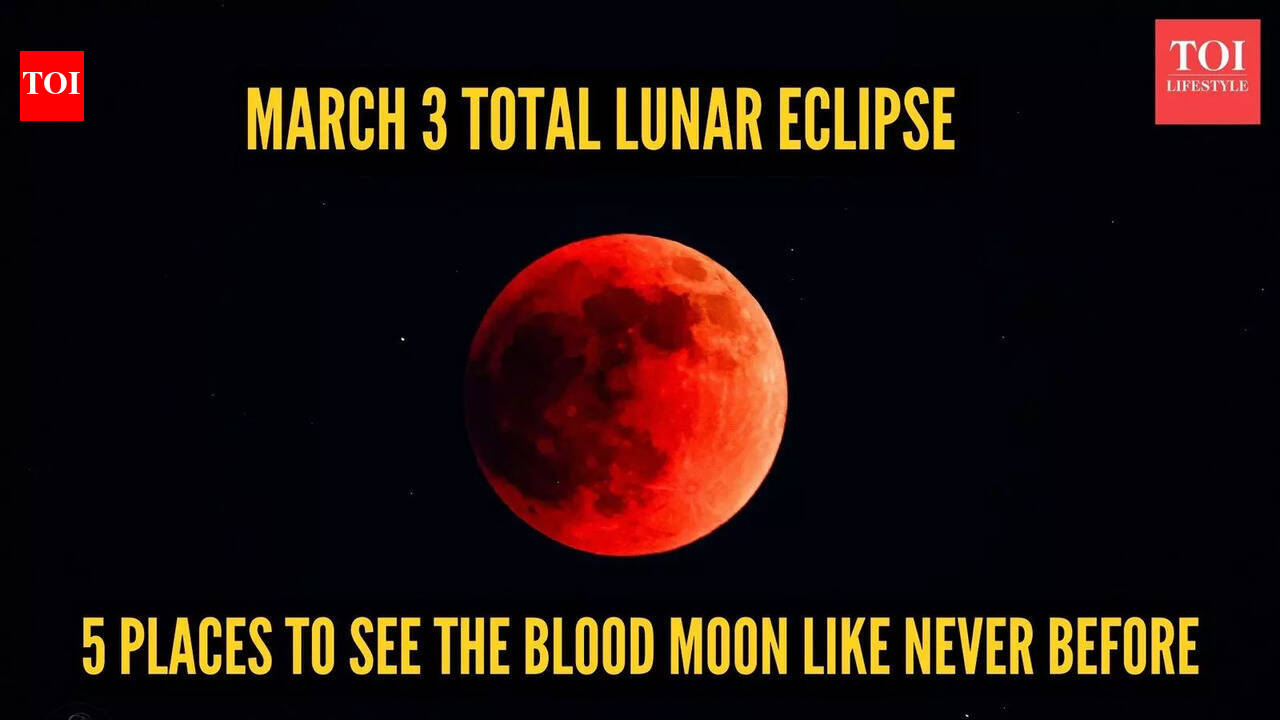
Here’s something exciting for everyone who loves looking at the sky! On March 3, 2026, the Moon will move completely into Earth’s shadow, making a total lunar eclipse. During this, the Moon will look deep red, which is why people call it a…

A virtual forearm can bend in a blink. It can also take its time, easing toward a target as if it is thinking about the move.
In a new virtual reality study, both extremes felt wrong.
When a prosthetic arm moves on its own, speed turns out to be…
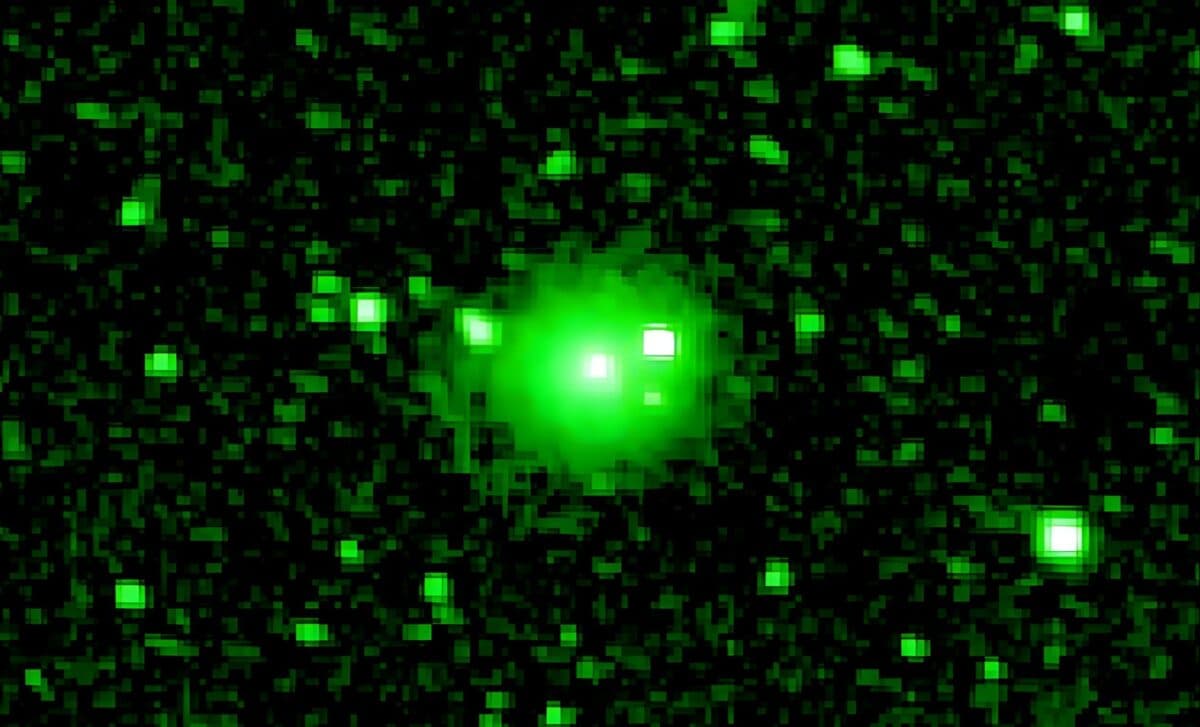
NASA’s SPHEREx space telescope has observed interstellar comet 3I/ATLAS flaring dramatically months after its closest pass by the Sun. Only the third confirmed interstellar object ever detected in our solar system, 3I/ATLAS is offering…