As oceans warm, many marine species are being squeezed into shrinking habitats, with coastlines cutting off their paths to cooler, safer waters.
When that escape fails, the losses spread beyond wildlife – disrupting food supplies, livelihoods,…

As oceans warm, many marine species are being squeezed into shrinking habitats, with coastlines cutting off their paths to cooler, safer waters.
When that escape fails, the losses spread beyond wildlife – disrupting food supplies, livelihoods,…

Images available for use with credit can be found here.
Newswise — Woods Hole, Mass. (January 20, 2026) — Researchers at the Woods Hole Oceanographic Institution (WHOI), in collaboration with the University of the…

Recfishwest and Woodside Energy have installed the Dampier Artificial Reef – a new, purpose-built artificial reef designed to support local fishers and enhance marine biodiversity off the Western Australian coast.
The Dampier…

The new infrared image from the NASA/ESA/CSA James Webb Space Telescope reveals the complex structure of gas and dust shed by a white dwarf in the center of the Helix Nebula.
This Webb image shows a portion of the Helix Nebula. Image credit:…

The Helix Nebula is one of the most well-known and commonly photographed planetary nebulae because it resembles the “Eye of Sauron.” It is also one of the closest bright nebulae to…

Deep beneath the surface of distant exoplanets known as super-earths, oceans of molten rock may be doing something extraordinary: powering magnetic fields strong enough to shield entire planets from dangerous cosmic radiation and other…

NASA will observe its annual Day of Remembrance on Thursday, Jan. 22, which includes commemorating the crews of Apollo 1 and the space shuttles Challenger and Columbia. The event is traditionally held every year on the fourth Thursday of…
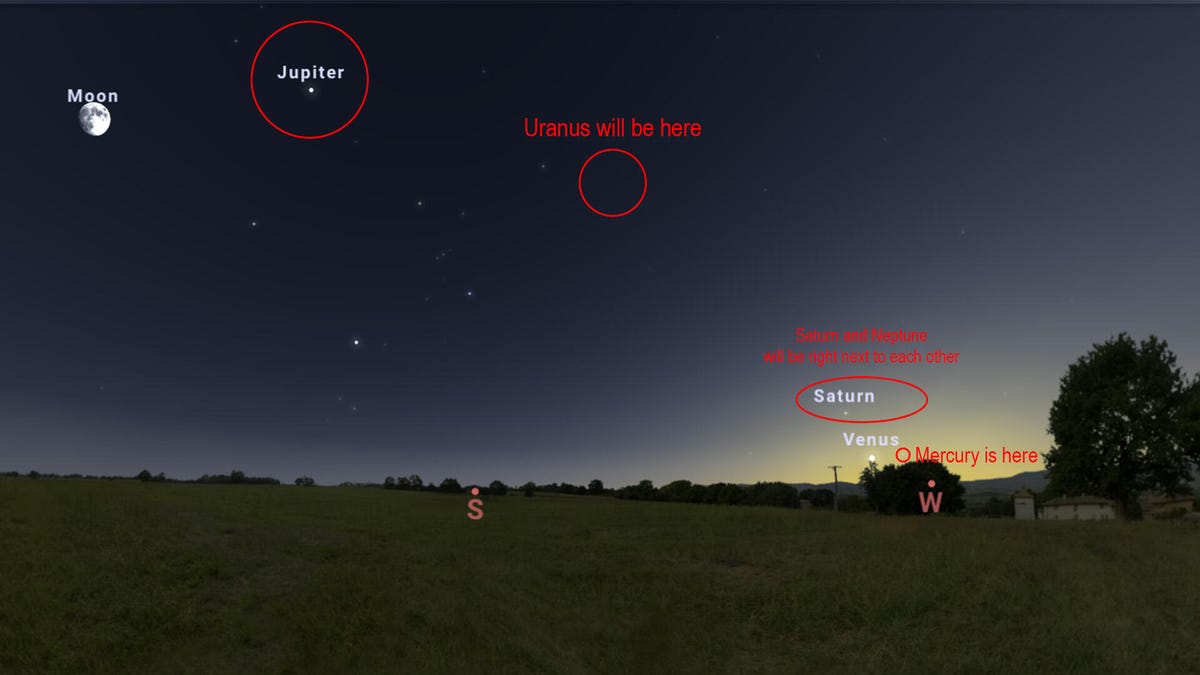
Skygazers, mark your calendars because one of the coolest celestial events is coming around again toward the end of February. Six planets will be visible in the night sky at the same time for a couple of weeks. This phenomenon is known as a…
New research challenges conventional wisdom by demonstrating that mid-ocean ridges and continental rifts, not volcanic eruptions, played the central role in atmospheric carbon swings and long-term climate shifts throughout Earth’s geological…