In contemporary China, an increasing number of names draw inspiration from Chinese history and classical texts. From high-tech products to newborns, names offer a window into the nation’s rich cultural heritage and ongoing…
Category: 7. Science
-

Our Moon’s 4-Billion-Year Impact Record Suggests Meteorites Didn’t Supply Earth’s Water
High-precision oxygen isotopes in Apollo lunar soils reveal a persistent impactor fingerprint, showing that impacts contributed only a tiny fraction of Earth’s water
WASHINGTON and…
Continue Reading
-
Researchers Use South Pole Telescope to Detect Energetic Stellar Flares Near the Center of the Milky Way | NCSA | National Center for Supercomputing Applications | Office of the Vice Chancellor of Research and Innovation
The universe is vast, but astronomers don’t have to look too far to find something genuinely new. Researchers at the Center for AstroPhysical Surveys (CAPS) used the South Pole Telescope to probe one of the most complex…
Continue Reading
-

Scripps Scientists in Antarctica Studying Retreating Glaciers, Cancer-Fighting Microbes and More
The GASP program grew out of an earlier collaboration between Scripps, Argentina’s Universidad Nacional de la Plata, and Antarctic tour operators called FjordPhyto, a citizen science program, in which tour guests assist trained…
Continue Reading
-

Walking sharks break the rules of reproductive energy costs
Sharks, rays, skates, and chimaeras are slow to reproduce, and scientists believe the process takes a lot of energy. But no one really knows how much, because it’s difficult to measure without harming the animals.
To learn more, researchers…
Continue Reading
-

What NASA Learned From Launching Artemis I
Episode description:
In 2022, NASA launched Artemis I, an uncrewed test flight of the rocket and spacecraft that will send humans to the Moon. Go inside Firing Room 1—the nerve center for Artemis launches—and hear from the engineers who…
Continue Reading
-
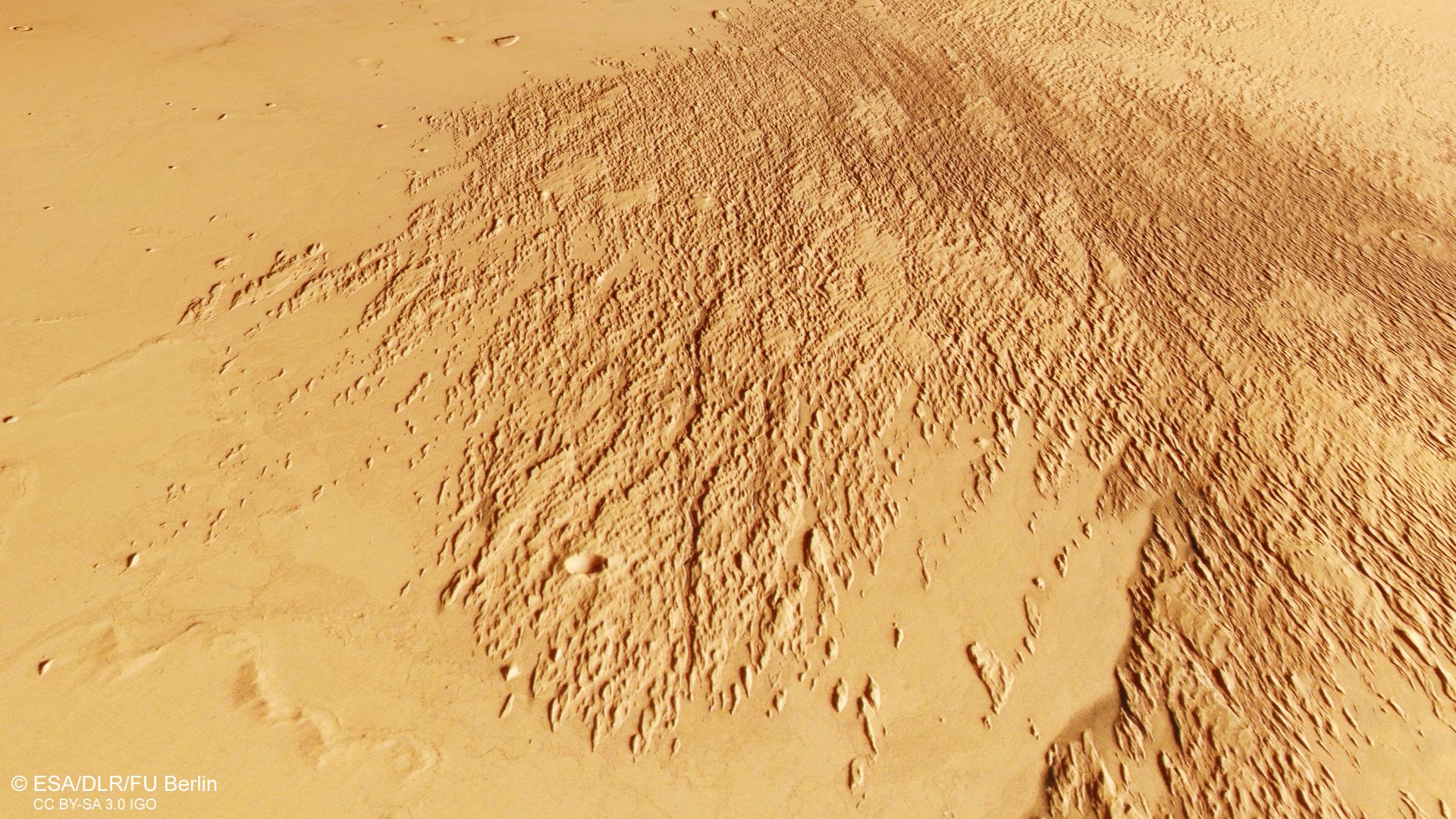
Mars orbiter sees odd etchings in the sand photo of the day for Jan. 20, 2025
Mars is famous for its volcanoes, canyons and ancient river valleys, but some of its most active geology happens in slow motion, powered by air. Over time, strong gusts can loft sand grains that ping and scrape at exposed surfaces, gradually…
Continue Reading
-

James Webb catches an exoplanet losing its atmosphere in real time
Astronomers from the University of Geneva (UNIGE), the National Centre of Competence in Research PlanetS, and the Trottier Institute for Research on Exoplanets (IREx) at the University of Montreal (UdeM) have made a major breakthrough using the…
Continue Reading
-
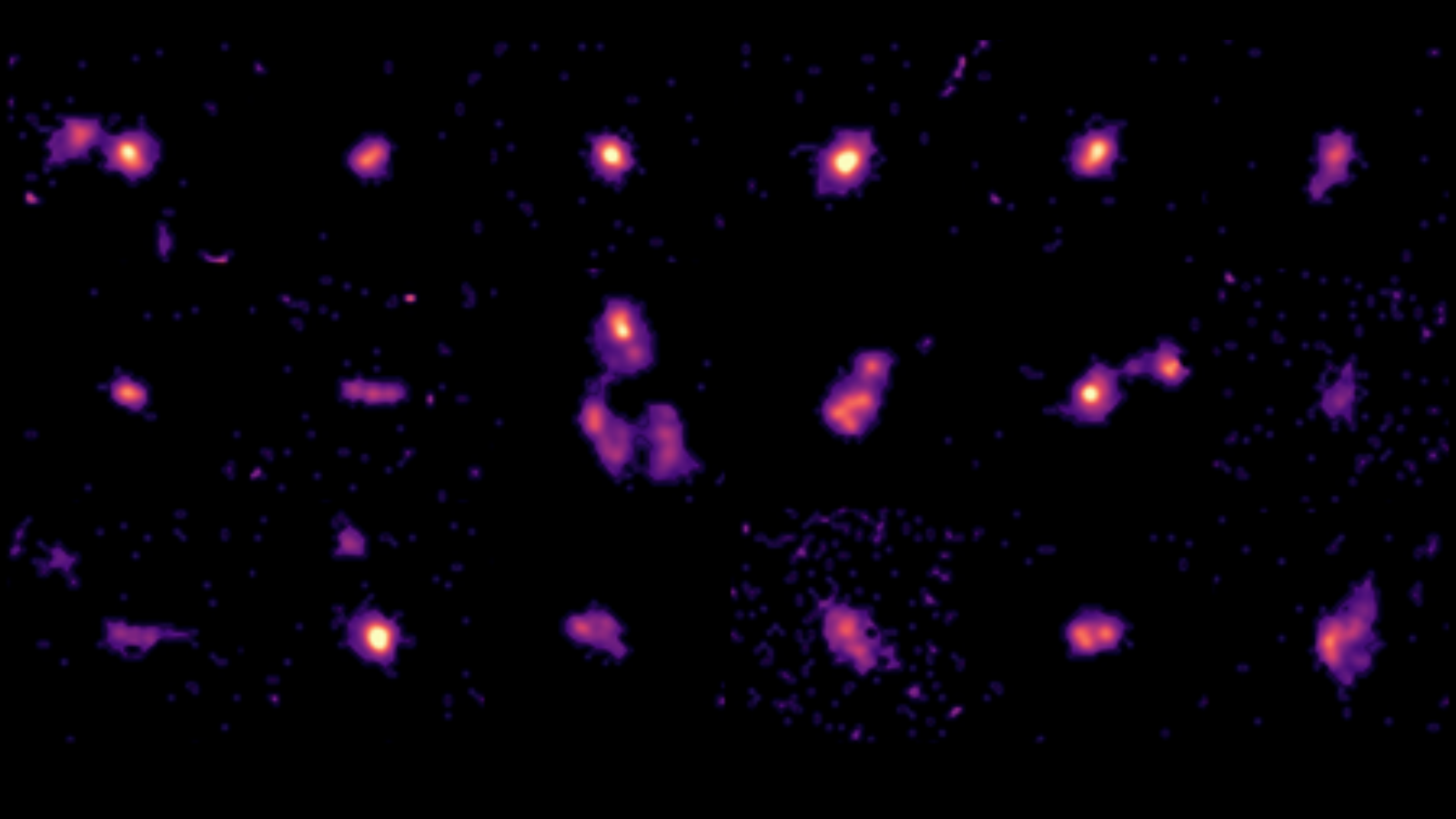
James Webb Space Telescope discovers young galaxies age rapidly: ‘It’s like seeing 2-year-old children act like teenagers’
Astronomers have obtained their most detailed look yet at young galaxies in the early universe using the James Webb Space Telescope, Hubble Space Telescope and Atacama Large Millimeter/submillimeter Array. The conclusion? These cosmic…
Continue Reading
-
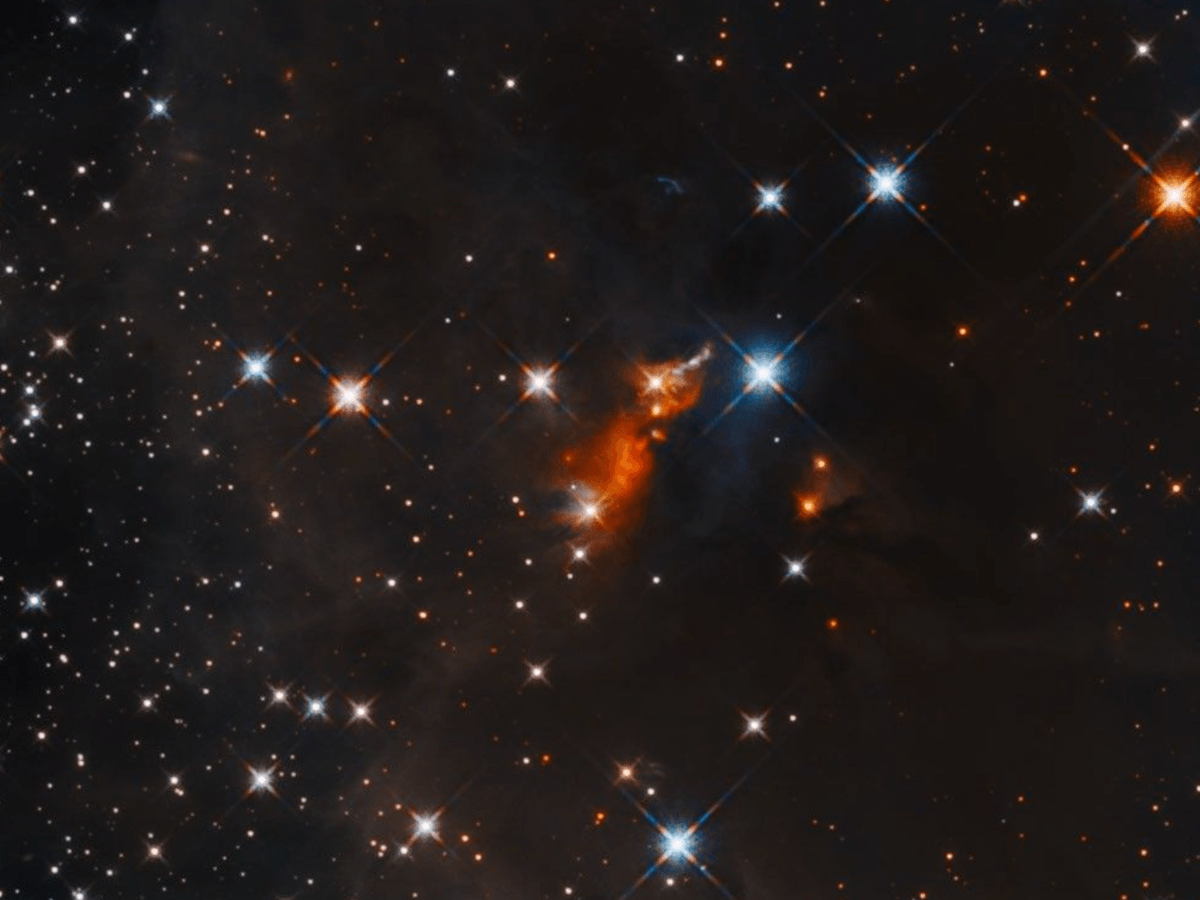
NASA’s Hubble Telescope captures striking images of developing stars
The Hubble Telescope took snapshots of the infant stars, in an effort to understand how larger stars are formed.
As part of the Sofia Massive Star Formation Survey, which is investigating how massive stars,…
Continue Reading