Dark matter may not have been ‘cold’ in the earliest moments after the Big Bang, as long believed; instead, new research from the University of Minnesota Twin Cities and the Universit’e Paris-Saclay suggests dark matter particles could…
Category: 7. Science
-

Damaged DSN antenna out of service until May
WASHINGTON — A key antenna in NASA’s Deep Space Network (DSN) that was damaged last fall is expected to remain offline until May, before being taken out of service again later this year for major upgrades.
The DSS-14 antenna, a…
Continue Reading
-
Walking sharks break biology reproduction rules – Phys.org
- Walking sharks break biology reproduction rules Phys.org
- Walking sharks breaking biology reproduction rules James Cook University
- VIDEO: These epaulette sharks are breaking the rules of biology Australian Broadcasting Corporation
- Walking Sharks…
Continue Reading
-
How a Global Freeze 445 Million Years Ago Changed Life Forever – SciTechDaily
- How a Global Freeze 445 Million Years Ago Changed Life Forever SciTechDaily
- Tiny fossils reveal ancient ecosystems that revived Earth’s oceans IOL
- UCT study uncovers resilient fossil ecosystem that revived ocean life after mass extinction Cape…
Continue Reading
-

An armada of 6,500 Elite Dangerous players just embarked on a three-month expedition to explore the Milky Way, and there’s still time to join them
Humanity is still in the early stages of exploring the cosmos, with Artemis 2 preparing to take us back to the moon and then onwards to Mars. But if NASA’s progress is a little pedestrian for you, then “Elite Dangerous” is the game for…
Continue Reading
-
Studying bubbles from the early Universe: an efficient matched filter approach
Title: Probing ionized bubbles around luminous sources during reionization with SKA 21-cm observations
Authors: Arnab Mishra, Kanan K. Datta, Chandra Shekhar Murmu, Samir Choudhuri, Iffat Nasreen, and Snehasish Saha
First author…
Continue Reading
-

Improving High-Resolution Mass Spectrometry Platforms
At the Winter Conference on Plasma Spectrochemistry, which took place in Tucson, Arizona, Spectroscopy sat down with Ken Marcus, a Robert Adger Bowen Professor of Chemistry at Clemson University, to talk about his research (1–2).
Marcus is an…
Continue Reading
-
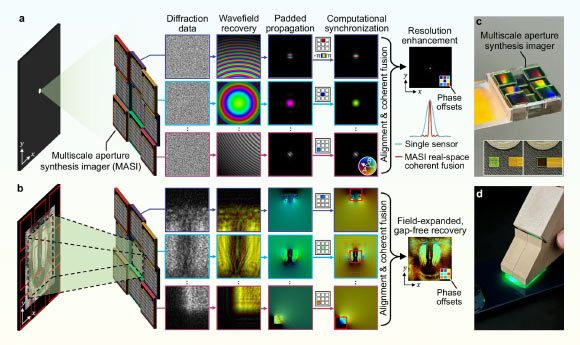
New Sensor Rewrites Rules of Optical Imaging
Inspired by a technique that allowed astronomers to image a black hole, scientists at the University of Connecticut developed a lens-free image sensor that achieves sub-micron 3D resolution, promising to transform fields from forensics to…
Continue Reading
-

Ancient Warming, CO2 Spurred Vast Fires, Erosion
56 million years ago, the Earth was already warm. ‘As a result, there was a lot of vegetation, even at high latitudes. That means that a lot of carbon was stored in, for example, vast coniferous forests.’ Biologist Mei Nelissen is…
Continue Reading
-

What the first medical evacuation from the International Space Station tells us about healthcare in space
This article was originally published at The Conversation. The publication contributed the article to Space.com’s Expert Voices: Op-Ed & Insights.
For the first time in 25 years of continuous crewed operations, an astronaut has been medically…
Continue Reading
