Scientists have produced a clearer picture of the landscape buried under Antarctica’s ice, revealing rigid valleys, channels, and other features that were previously hard to detect.
Those hidden shapes matter because they steer ice flow and…
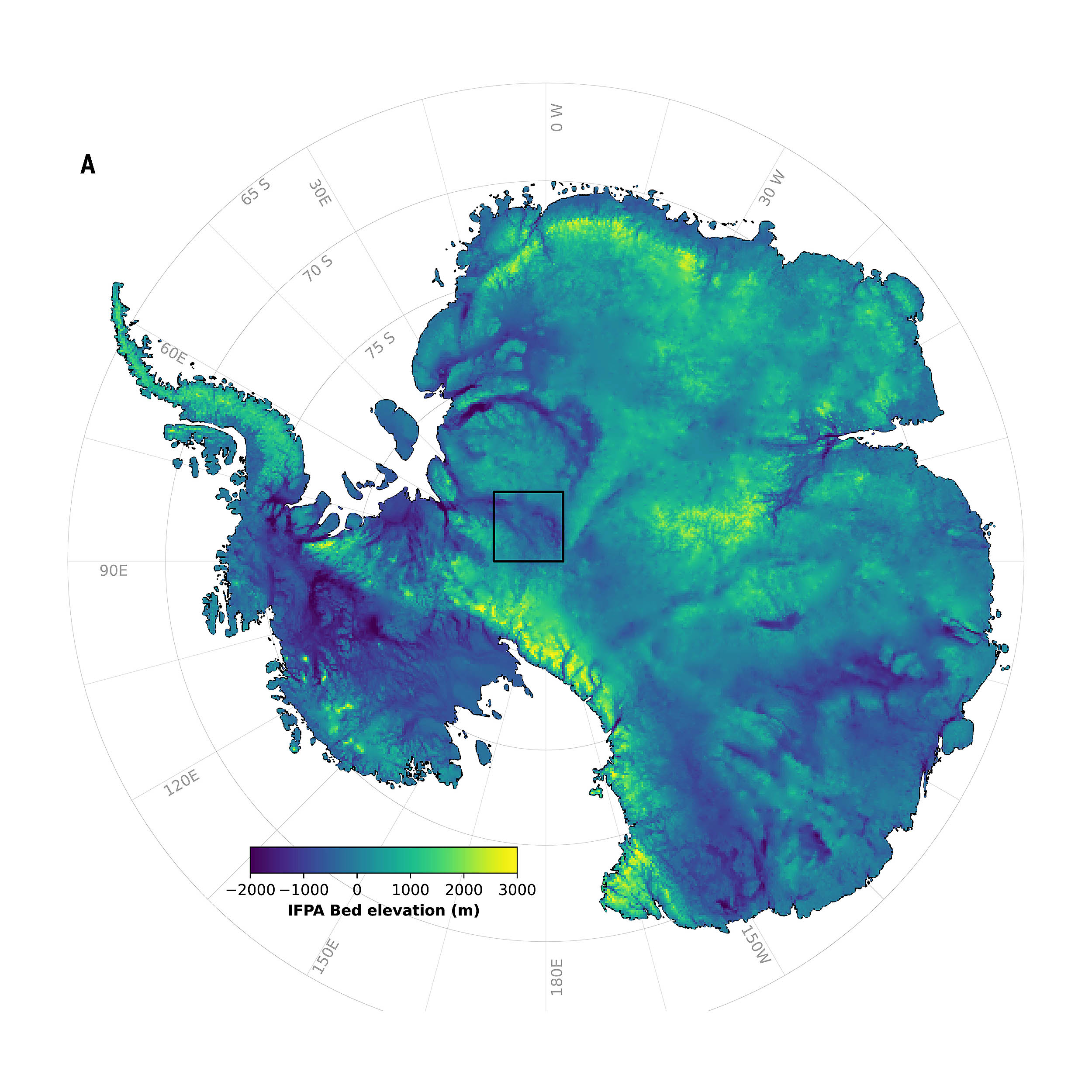
Scientists have produced a clearer picture of the landscape buried under Antarctica’s ice, revealing rigid valleys, channels, and other features that were previously hard to detect.
Those hidden shapes matter because they steer ice flow and…

The comet, initially identified as 6AC4721 and now set to be officially named “MAPS” after its discoverers, could be visible to the naked eye by late March, ahead of its closest approach to the Sun around 4 April.
The discovery was made by…

The southernmost extent of mainland Canada, along the northern shore of Lake Erie, lies at about the same latitude as Des Moines, Iowa. Though not a “breadbasket” like the grain-producing machine that is the U.S. Midwest, this part of…
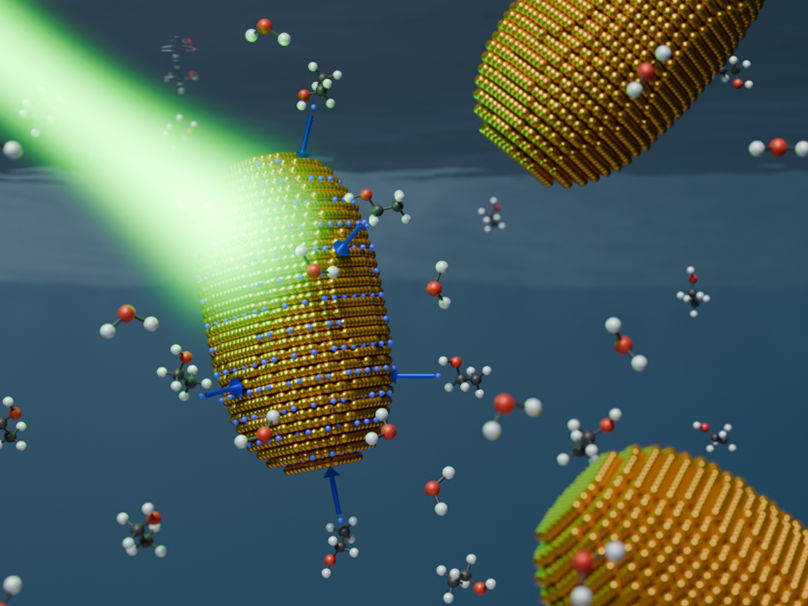
For the first time, a research group at the University of Potsdam has succeeded in tracking and quantifying the light-induced accumulation of charge on gold nanorods in real time. A new physical model of this process,…
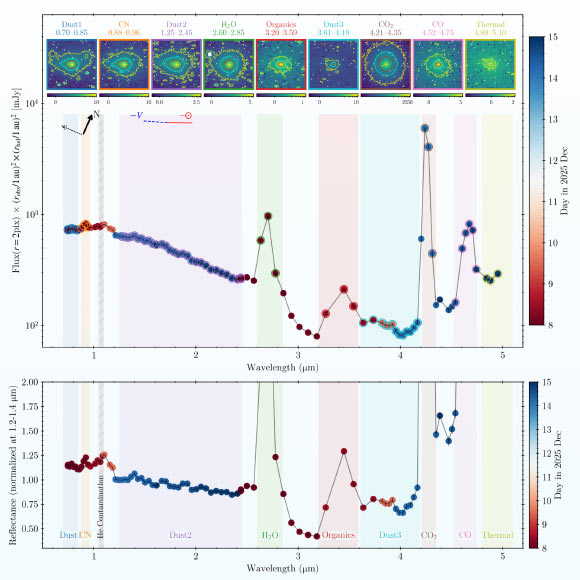
According to new observations by NASA’s SPHEREx mission, the interstellar object 3I/ATLAS has dramatically changed its behavior, developing the hallmarks of a fully active comet after a close encounter with the Sun.
The SPHEREx 0.75-5.0μm…
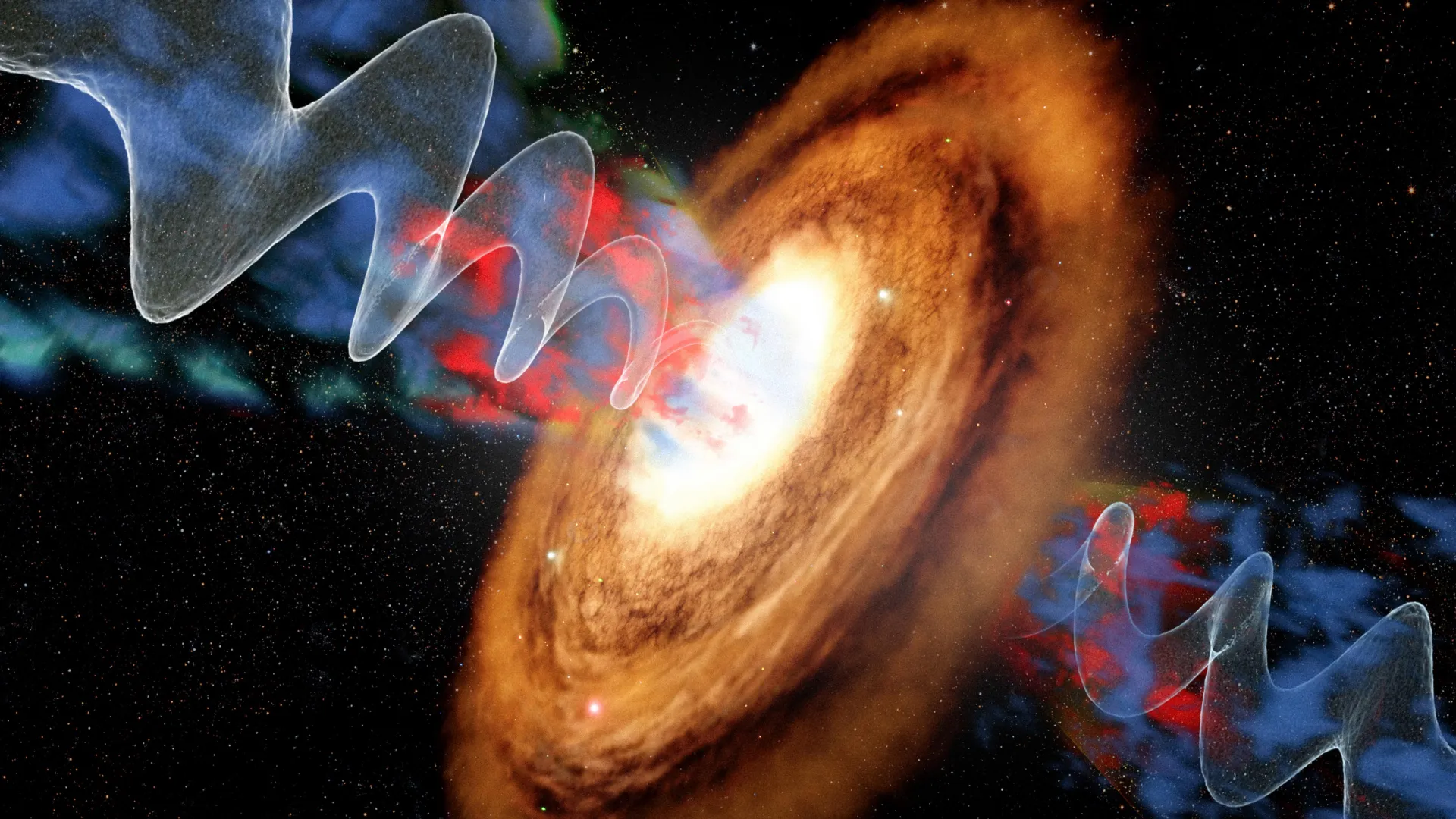
Some galaxies host an active galactic nucleus, an intensely bright region powered by a supermassive black hole that is actively pulling in surrounding matter. As gas and dust spiral toward the black hole, enormous amounts of energy can be…

A new study suggests Antarctic ice melt is being accelerated by a dangerous double force, with warming air and water working together to destabilize the continent’s massive ice sheets.
The study, published in the journal Nature…

On January 3, NASA closed the library at the Goddard Space Flight Center in Greenbelt, Maryland, the largest research library in the agency. The closure follows months of chaotic building shutdowns, laboratory dismantling and workforce reduction…

Researchers from IPHES-CERCA have contributed to a new study led by the National Research Center on Human Evolution (CENIEH) that challenges long-standing ideas about how early humans survived. Published in the journal Journal of Human Evolution,…