Pathogens and clinical specimens
CSTAdV-1 [5], Citrobacter freundii (Cf) LCJY-002 [13], Aeromonas veronii (Av) Av-202201, Aeromonas hydrophila (Ah) Ah-202401 and Morganella morganii (Mm) Mm-202407 were isolated and preserved in our laboratory. Soft-shelled turtle iridovirus (STIV), Trionyx sinensis hemorrhagic syndrome virus (TSHSV) were generously provided by Dr. Liu Hong of Shenzhen customs district, P.R. China, and Dr. Liu Li of Institute of hydrobiology, Zhejiang Academy of Agricultural sciences, P.R. China, respectively. Vibrio cholerae (Vc) was provided by the Nanchang City Center for Disease Control and Prevention, jiangxi Province, China. During April to June in 2024, a total of 107 clinical internal organ tissues (liver, kidney, spleen) from diseased Chinese soft-shelled turtles were collected from farms in Nanfeng County of Fuzhou city, Jiangxi Province, China. This county produces more than a half of the Chinese soft-shelled turtle eggs and seedlings in China [14]. The clinical features of these diseased Chinese soft-shelled turtles included: putrid Skin, hepatomegalia, intestinal bleeding, intestinal obstruction, parotiditis etc.
DNA/RNA Extraction and Generation of DNA Standard
Total DNA/RNA of all viruses was extracted from 100 μL of virus suspensions using the TaKaRa MiniBEST Viral RNA/DNA Extraction Kit (Beijing, China) according to the manufacturer’s instructions. The cDNA of TSHSV was synthesized using the TaKaRa PrimeScript™ 1 st strand cDNA Synthesis Kit (Beijing, China). Bacterial genomic DNA was extracted using the TaKaRa MiniBEST Bacteria Genomic DNA Extraction Kit (Beijing, China) according to the manufacturer’s instructions. Total DNA of 107 clinical internal organ tissues (liver, kidney, spleen) from diseased Chinese soft-shelled turtles was extracted from 200 μL of tissue suspensions using the Qiagen DNeasy Blood & Tissue Kit (Hilden, NRW, Germany) according to the manufacturer’s standardized instructions, the concentration and quality of DNA were tested using BioPhotometer Plus Nucleic acid/Protein analyzer (Eppendorf, Germany) after DNA extraction. The CSTAdV-1 DNA polymerase gene segments (3,324 bp, GenBank accession no. PQ083072) were synthesized by Sangon Biotech (Shanghai, China) and cloned into a pUC57 vector, designated as pUC57-CSTAdV. The pUC57-CSTAdV standard plasmid DNA was extracted using TIANGEN TIANprep Mini Plasmid Kit (Beijing, China) and then measured by BioPhotometer Plus Nucleic acid/Protein Analyzer (Eppendorf, Germany), The DNA copy number was calculated using the following equation: DNA copy number = (M × 6.02 × 1023 × 10–9)/(n × 660). The calculation result is 1.0 × 109 copies/μL. The pUC57-CSTAdV standard plasmid DNA was subsequently diluted tenfold with TE buffer. Both DNA samples were stored at −80 °C until use.
Design of the Primer and Probe of RF-RAA and qPCR
Based on the complete genome sequence of CSTAdV-1 (GenBank accession no. PQ083072), the DNA polymerase gene was selected for primer and probe design after comparative analysis of conserved regions across previously identified positive samples. In accordance with the guidelines provided by ZC Bioscience RAA kits (Hangzhou Zhongce Bio-Sci & Tech Co. Ltd., China), three forward primers, two reverse primers, and two probes were designed to identify the most efficient combination for RF-RAA assays. Additionally, forward and reverse primers and along with a probe were designed for qPCR analysis using Primer Premier 5.0 software. All designed primers and probes were subjected to BLAST analysis to verify their specificity. Subsequently, they were synthesized by Sangon Biotech (Shanghai, China). The sequences of the designed primers and probes were summarized in Table 1.
Optimization of reaction conditions for RF-RAA
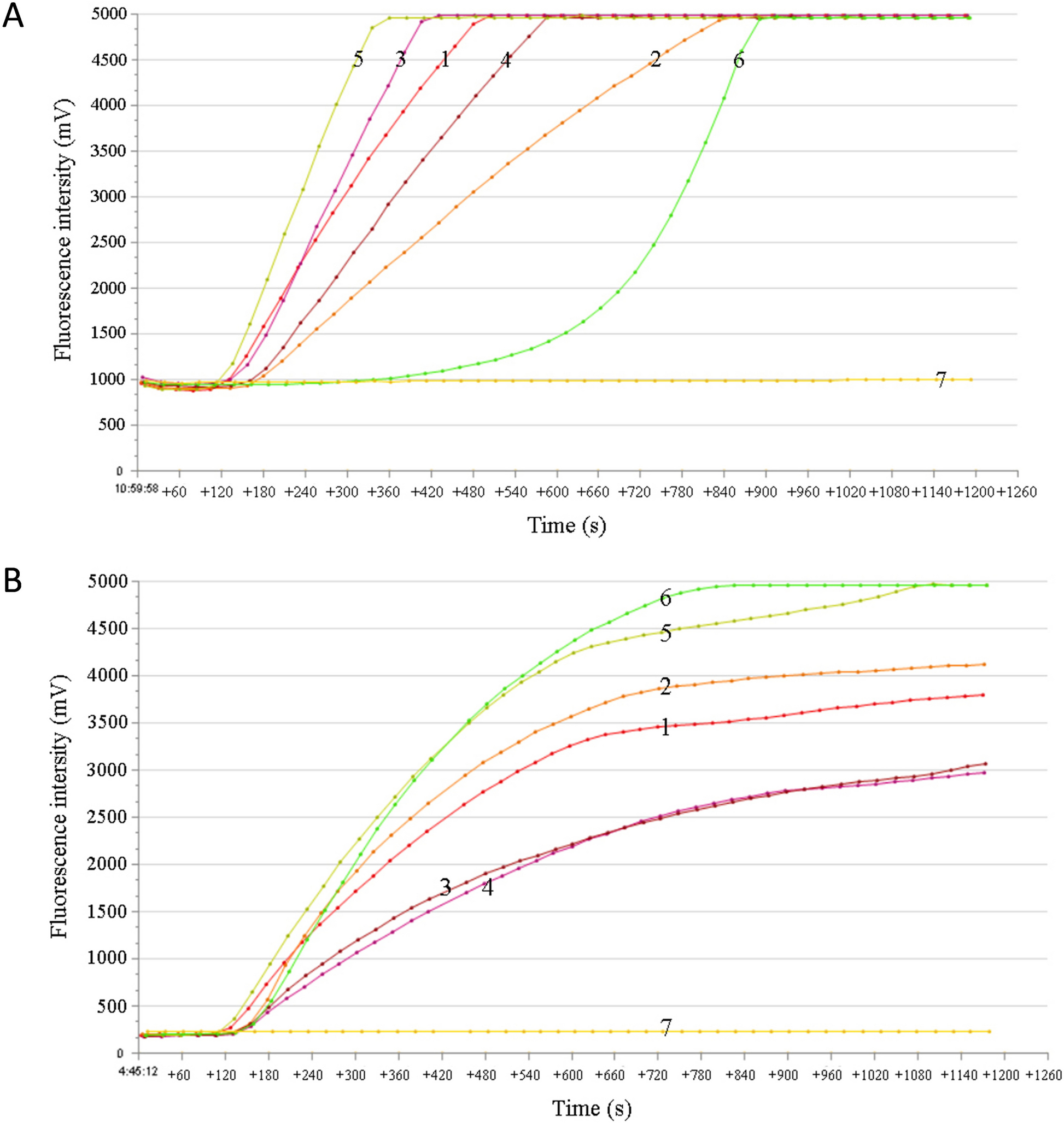
The probe-based CSTAdV RF-RAA assay was performed using a fluorescence detection device T16-ISO (TwistDx, United Kingdom) in a 50 μL reaction volume. The reaction mixture was prepared using the ZC Bioscience RAA exo kit (Hangzhou Zhongce Bio-Sci & Tech Co. Ltd., China), which included 25 μL of A buffer (rehydration buffer), 5 μL of template DNA, 2 μL each of forward and reverse primers (10 μM, Sangon Biotech, China), 0.6 μL of RAA exo probe (10 μM, Sangon Biotech, China), 12.9 μL of ddH2O and 2.5 μL of B buffer (280 mM, magnesium acetate). The reaction time was carried out for 20 min. To determine the optimal reaction conditions, different combinations of primers and probes were tested to amplify 1.0 × 107 copies/μL of pUC57-CSTAdV DNA as the template, with three replicates for each condition. Additionally, the assay was performed at different incubation temperatures (35 °C, 37 °C, 39 °C, 40 °C, and 42 °C). Fluorescence intensity was measured using the FAM channel (excitation 470 nm and detection 520 nm) for 20 min.
Optimization of reaction conditions for qPCR
The TaqMan probe-based CSTAdV qPCR assay was performed on a Gentier 96E quantitative fluorescence instrument (Tianlong, China) in a 20 μL reaction volume using the TaKaRa Probe qPCR Mix kit (Beijing, China). Using 1.0 × 107 copies/μL of pUC57-CSTAdV DNA as the template, the annealing temperature was optimized by testing a gradient range of 50–62 °C. Additionally, different final concentrations of primers (0.1–0.8 μmol/L) and probe (0.1–0.5 μmol/L) were tested to determine the optimal combination that resulted in the lowest critical cycle number (Ct) and the highest relative fluorescence units (RFU).
Specificity of the RF-RAA and qPCR assays
The specificity of the developed CSTAdV RF-RAA and qPCR assays were evaluated by detecting a panel of pathogens, including CSTAdV-1, STIV, TSHSV, Cf, Av, Ah, Mm, and Vc. All detection assays were performed in triplicate, using pUC57- CSTAdV (1.0 × 107 copies/μL) as the positive control and ultrapure water as the negative control.
Sensitivity of the RF-RAA and qPCR assays
To evaluate the sensitivity of RF-RAA and qPCR assays, tenfold serial dilutions of the standard plasmid pUC57-CSTAdV DNA (ranging from 1.0 × 106 copies/μL to 1.0 copy/μL) were used as templates to determine the detection limits. All detection assays were performed in quintuplicate, with ultrapure water serving as the negative control. Standard curves for the qPCR assay were generated based on the concentration gradient of the diluted plasmid and Ct values.
Repeatability of the RF-RAA and qPCR assays
The repeatability of the CSTAdV RF-RAA and qPCR assays was evaluated using pUC57-CSTAdV plasmid standards at concentrations of 1.0 × 106 copies/μL and 1.0 × 103 copies/μL as templates. Intra-group repeatability tests were performed with three replicates for each concentration. Additionally, three independent inter-group repeatability tests were conducted under identical conditions at 7-day intervals to further assess the repeatability of the methods.
Practical application of the clinical samples
The practicality of both the CSTAdV RF-RAA and qPCR assays was evaluated using 107 archived DNA samples extracted from clinical internal organ tissues of diseased turtles. The results were compared with those obtained from the adenoviral consensus nested PCR assay, with both the reaction system and amplification protocol of the nested PCR being strictly implemented according to the referenced methodology [15]. The positive control was pUC57-CSTAdV plasmid DNA (1.0 × 107 copies/μL), the negative control was DNA from healthy Chinese soft-shelled turtles, and the blank controls was ultrapure water. Each assay was performed in duplicate. Diagnostic sensitivity (DSe) and diagnostic specificity (DSp) were calculated following the guidelines provided in the 2023 edition of the Manual of Diagnostic Tests for Aquatic Animals (WOAH).