Participants
The diagnostic criteria referred to international experts’ consensus (2018 version). The clinical manifestations of BWS were categorized into cardinal features and suggestive features. The BWS score was 2 points for each cardinal feature and 1 point for each suggestive feature. Inclusion criteria were a clinical diagnosis of BWS with a score of ≥ 4 points or a definite molecular diagnosis of BWS with a score of ≥ 2 and < 4 points. Exclusion criteria were incomplete clinical data, refusal to participate by the guardians, and combination of BWS with other hereditary diseases.
Phenotyping
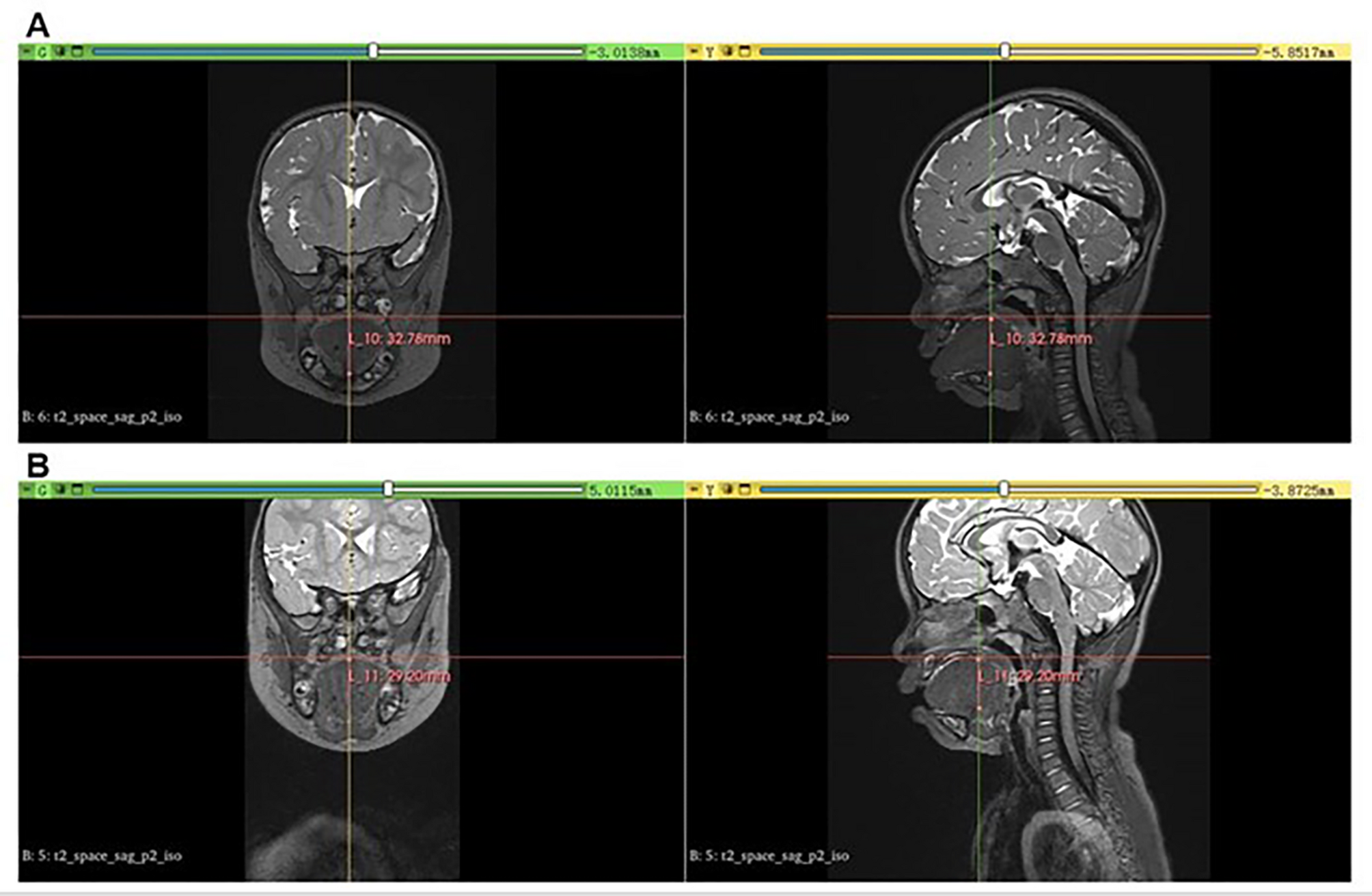
Definitions of clinical manifestations of BWS refer to previous literature. Macroglossia was defined as a tongue that protrudes beyond the teeth at rest or a tongue margin with teeth marks when the mouth is slightly opened [11]. The BWS children with macroglossia underwent MRI scans preoperatively and postoperatively to demonstrate the size of the tongue. We conducted measurements using 3D Slicer software to visualize the evaluation. Figure 1 shows MRI scanning and measurements using 3D Slicer software of the macroglossia in BWS children. Lateralized overgrowth (LO) was defined as asymmetric overgrowth of one or more limbs and/or the face due to abnormal cell proliferation without any other diagnosis, with a visible asymmetry and/or measured asymmetric difference of up to 5% [12]. Isolated lateralized overgrowth (ILO) was defined as lateralized overgrowth lacking known malformations and developmental and morphological abnormalities [13]. Transient hypoglycemia in neonates was defined as a low blood glucose level lasting < 1 week; Hyperinsulinism was defined as prolonged hypoglycemia in the context of elevated insulin levels that last beyond one week and/or require escalated treatment [4].
MRI scanning and measurements using 3D Slicer software of the macroglossia in BWS children in coronal and sagittal positions. Head and neck MRI scanning shows severe macroglossia protruding preoperatively (A) in the oral cavity postoperatively (B)
(Epi)genotyping
MS-MLPA is the first-line test for the molecular diagnosis of BWS. All molecular tests were performed using peripheral blood. Genomic DNA was extracted from peripheral blood using the QIAamp DNA Mini kit (Qiagen, Hilden, Germany). MS-MLPA assays were performed using the SALSA MLPA ME030-C1 BWS/RSS kit (MRC-Holland, Amsterdam, the Netherlands) to detect copy number variation (gene duplications/deletions) in the 11p15 region (IC1 and IC2) and methylation status of 8 CpG islands. MS-MLPA included a 5-step process, denaturation and hybridization, ligation, PCR amplification, fragment isolation, and data analysis performed MS-MLPA. Genomic DNA (200 ng) was denatured and hybridized by mixing the probes at 60 °C for 16 h. The samples were divided equally into two portions; one was treated with ligase-65 ligase to detect copy number variation, and the other was digested with HhaI restriction enzyme before treatment with ligase to detect the methylation status of the 11p15.5 region. PCR was then performed using fluorescently labeled probe primers, including IC1 and IC2. Amplified products were separated on an ABI 3500Dx Genetic Analyzer (Applied Biosystems, Foster City, CA, USA), and the results were analyzed using Coffalyser Net software (MRC-Holland, Amsterdam, Netherlands). GOM was defined as methylation 20% higher than normal controls, LOM was defined as methylation 20% lower than normal controls, and the methylation level of normal controls was 50%. The presence of both IC1 GOM and IC2 LOM was diagnostic of pUPD11. Heterozygous deletions (or duplications) were defined as a 35%-50% decrease (or increase) in the relative peak area of the amplification product [14, 15]. To investigate potential variants in the CDKN1C gene, next-generation sequencing (NGS) was performed on genomic DNA extracted from patient samples. DNA was isolated using a standard genomic DNA extraction protocol and quantified to ensure sufficient input for sequencing. The CDKN1C gene was targeted for sequencing using custom-designed probes or a commercially available gene panel. Sequencing libraries were prepared according to the manufacturer’s protocol, and sequencing was carried out on an Illumina platform (or specify the platform used). Chromosomal abnormalities analysis was performed using Karyotyping. Peripheral blood samples (or other tissue types) were collected from patients, and lymphocytes were cultured using standard techniques. Chromosomes were analyzed under a light microscope at high magnification, and the chromosome number and structure were assessed for any abnormalities, including duplications, deletions, translocations, or inversions.
(Epi)genotype-phenotype correlations
In this study, the clinical data of the BWS children were collected, including name, gender, age, gestational age at birth, birth weight, clinical manifestations, clinical scores, molecular diagnosis, and MRI imaging data. (Epi)genotype-phenotype correlations were analyzed in BWS patients with molecular diagnoses of IC2 LOM, IC1 GOM, and pUPD11. To obtain reliable (epi)genotype-phenotype correlation results, BWS cases with only a clinically confirmed diagnosis but lacking a molecular diagnosis and cases with negative MS-MLPA test results were not included in the analysis. Cases with chromosome 11p15 abnormalities and CDKN1C mutations were also excluded due to the disproportionately limited cases.
Statistical analysis
Descriptive analysis demonstrated the characteristics, clinical manifestations, and molecular abnormalities of the BWS children. Quantitative data that satisfy normal distribution are presented as mean ± standard deviation (x̅±s); quantitative data that do not satisfy normal distribution are presented as percentage (n%) and median. Continuous variables were compared using t-tests; (Epi)genotype-phenotype difference analyses between the IC2 LOM, IC1 GOM, and pUPD11 groups were compared using 3 × 2 χ2 tests or Fisher’s exact tests. All statistical analyses were performed using SPSS 26.0, and p < 0.05 was considered statistically significant.