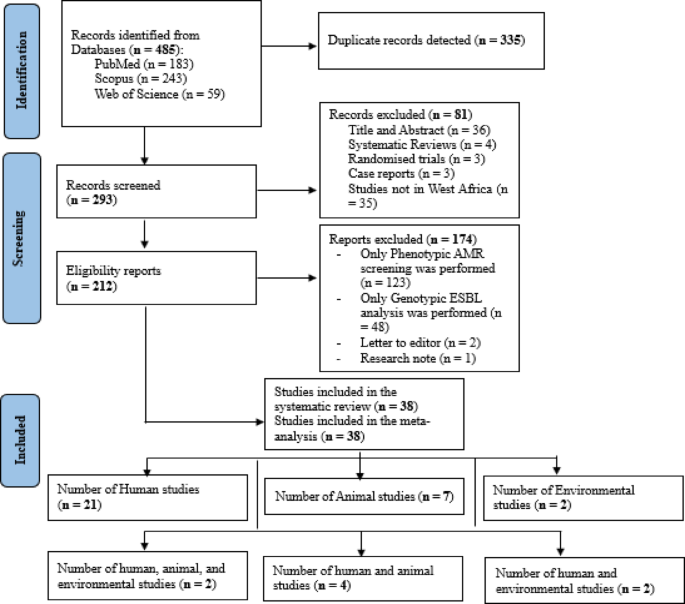
Search and screening results
A total of 485 studies were retrieved after the initial search across three databases, including 183 from PubMed, 243 from Scopus, and 59 from Web of Science. After 335 duplicate studies were detected and resolved, titles and abstracts of 293 articles were screened based on the eligibility criteria. A total of 212 studies which were initially considered eligible and were subjected to full-text evaluation. After full-text examination, 38 articles were eligible for inclusion [humans (n = 21), animals (n = 7), environment (n = 2), Interdisciplinary studies (n = 8)] (Fig. 1).
PRISMA flowchart of study selection.
Characteristics of included studies
All included studies were published between 2011 and 2024, with most of them (63.2%) having been conducted between 2015 and 2019. Studies that met the inclusion criteria were from only 9 out of 16 countries in West Africa. Nigeria had the highest number of individual studies (n = 17), followed by Ghana (n = 13), Senegal (n = 3), and Burkina Faso (n = 2). The remaining countries, Guinea-Bissau, Guinea, Niger, Gambia, and Benin, had one study each (Fig. 2) (Supplementary data 1; Table 3).
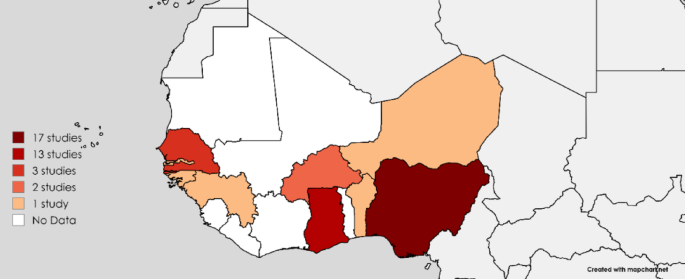
Distribution of studies conducted in West Africa.
The number of confirmed ESBL-positive isolates across the studies ranged from 1 to 1937 and comprised 25 different bacterial isolates across the nine West African countries, encompassing various populations, settings, and sample types (Fig. 3) (Supplementary data 2; Table 1).
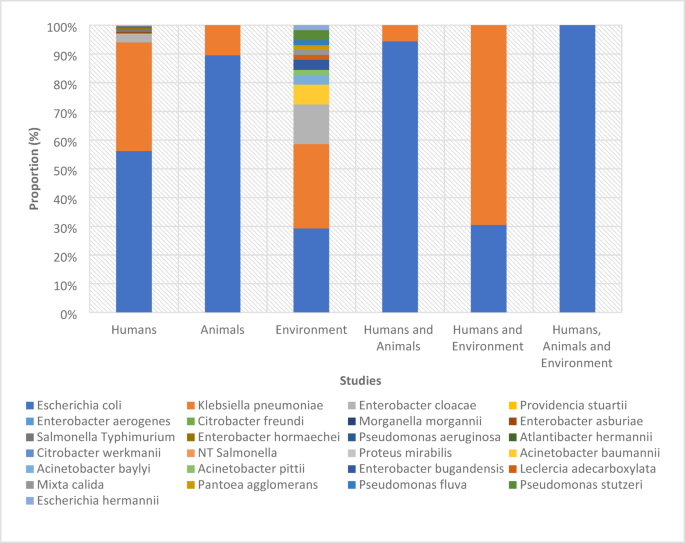
ESBL positive bacterial isolates from different studies.
A majority of the studies were conducted on humans (76.3%, n = 29), followed by animals (34.3%, n = 13) and the environment (15.6%, n = 6) (Supplementary data 1; Table 3). Most studies isolated ESBL-positive bacteria from hospital-based settings (68.2%, n = 24), followed by communities (15.8%, n = 6), farms (10.5%, n = 4), and markets (2.6%, n = 1). Some isolated the bacteria from a combination of sources, such as markets and farms (7.9%, n = 3) and communities and hospitals (5.3%, n = 2). Most studies isolated the bacteria from two major sample types: clinical specimens (39.5%, n = 15) and faeces (31.6%, n = 12) (Supplementary data 1; Table 3).
Among the 25 ESBL-positive isolates, 18 were identified as Enterobacteriaceae and 7 were classified as non-Enterobacteriaceae (Supplementary data 2; Table 1).
The most commonly used ESBL diagnostic methods were EUCAST disk diffusion (18 studies), Double-Disk Synergy Test (DDST) (18 studies), Whole Genome Sequencing (WGS) (30 studies), and Multilocus Sequence Typing (MLST) (34 studies) (Supplementary data 1; Table 3).
ESBL-producing bacterial species across human, animal, and environmental sources in West Africa
E. coli and K. pneumoniae accounted for 64% and 31% of the ESBL-positive isolates, respectively. Other Enterobacteriaceae species collectively comprised 4% of the isolates, whereas non-Enterobacteriaceae constituted 1% of the total isolates (Supplementary data 2; Table 1A).
ESBL genes and clones
A total of 171 ESBL genes were identified, with blaCTX-M being the most predominant gene (70.8%) across humans, animals, and the environment. The most frequent variant, blaCTX-M-15 (31.6%), was detected in all three sources, with greater occurrence in humans (21.1%) than in animals (7.6%) or the environment (2.9%) (Fig. 4) (Supplementary data 2; Table 2–10).
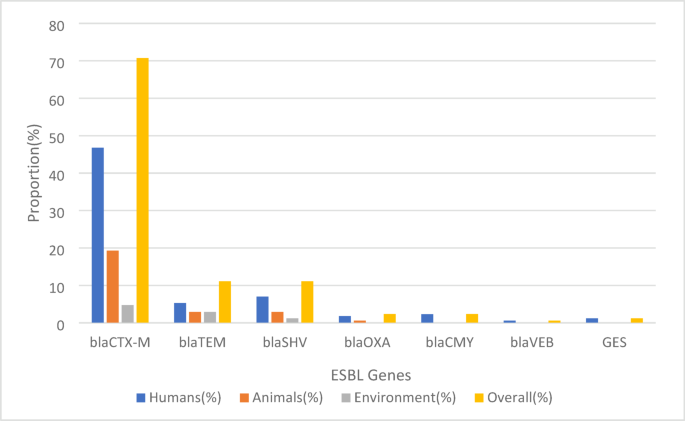
Prevalence of ESBL genes across human, animal, and environmental sources.
Multilocus sequence typing (MLST) was performed on 7 of 25 isolates, identifying 153 unique sequence types (STs) (Supplementary data 2; Table 14). Among these, Nigeria accounted for 133 clones from 5 isolates, with ST10, ST131, and ST410 being the most frequently observed. Ghana contributed 139 clones from 3 isolates, predominantly featuring ST131 and ST617, whereas 64 clones were identified from 4 isolates in other countries (Supplementary data 2; Table 11–13).
In terms of species-specific observations, E. coli exhibited considerable differences in sequence types among the different sources. In humans, ST131, ST10, ST410, and ST617 were prominent, whereas animals predominantly exhibited ST48, ST10, and ST38. Environmental samples were marked by the prevalence of ST10, ST58, and ST155. For K. pneumoniae, the predominant human STs were ST17, ST36, ST530, ST15, and ST14. In animals, ST307 was observed, and ST152 was most frequently observed in the environment. Non-Enterobacteriaceae displayed notable source-specific trends, with Pseudomonas aeruginosa ST1469 and ST235 linked to human samples and Acinetobacter baumannii ST109, ST1, ST78, ST132, and ST1136 linked to environmental samples (Supplementary data 2; Table 11–13).
Clonal distribution analysis showed that 71 clones were unique to humans, 27 to animals, and 14 to the environment. The number of shared clones between humans and animals was 25, between humans and the environment was 4, and between animals and the environment was 5. Notably, seven clones including ST10, ST410, ST58, ST155, ST4684, ST2178, and ST37 were detected predominantly in E.coli across all three sources. ST37 was also observed in K. pneumoniae (Fig. 5) (Supplementary data 2; Tables 11–14; Fig. 1).
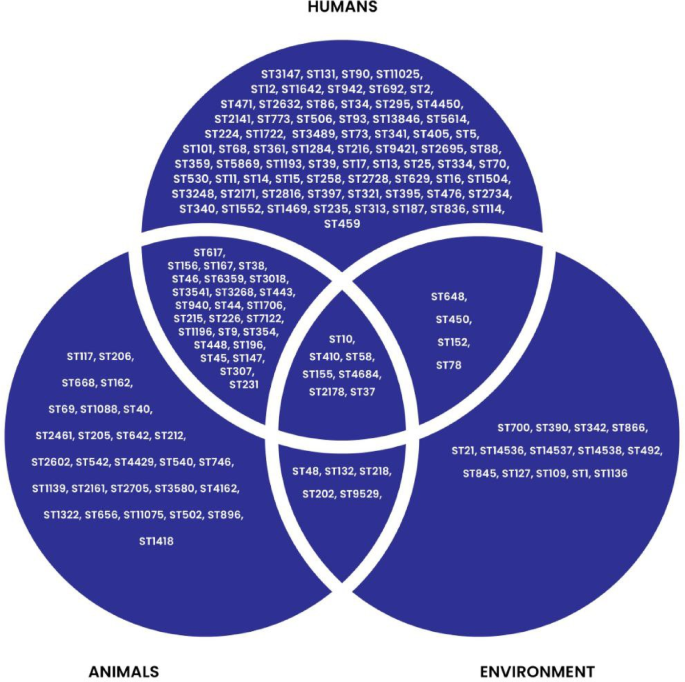
Clonal distribution across West Africa.
Potential transmission routes of ESBL clones
Data from 7 of 38 studies (18.4%) were analysed for possible transmission routes of ESBL clones across human, animal and environmental sources. These studies used a combination of sources to detect the prevalence of ESBL-producing bacteria in hospitals, markets, and farms (Supplementary data 1; Table 3). These studies included Humans-Animals, Humans-Environment, and Humans-Animals-Environment studies (Table 1). Although the studies did not clearly state the transmission routes, they acknowledged that because of the common ESBL clones detected, the transmission routes were possible.
Antimicrobial resistance patterns
An analysis of 24 studies covering 1215/1937 ESBL-producing bacterial isolates revealed significant resistance patterns against non-beta-lactam antibiotics (Supplementary data 1; Table 4) (Supplementary data 2; Table 15–15A). The results show that bacteria from humans are much more resistant than those from animals and the environment. Phenotypically, the bacteria were particularly resistant to commonly prescribed antibiotics such as ciprofloxacin (71%), gentamicin (58%), and trimethoprim-sulfamethoxazole (53%). Resistance to tobramycin (39%) and tetracycline/doxycycline (28%) was moderate, whereas nalidixic acid (17%), imipenem/meropenem (16.7%), and chloramphenicol (14.1%) showed lower resistance (Fig. 6).
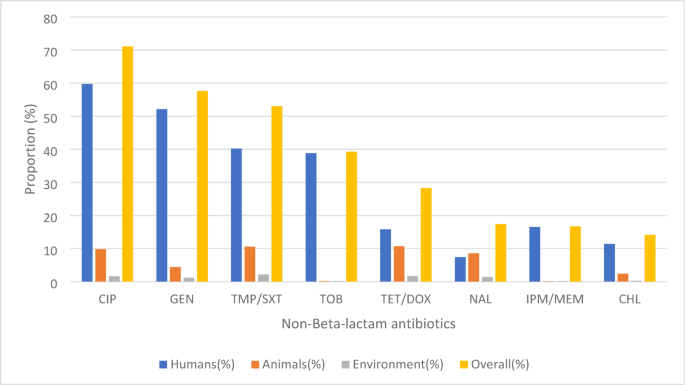
Antimicrobial resistance of ESBL bacterial isolates to non-beta-lactam agents.
Co-resistance genes
The ESBL-producing bacterial isolates also exhibited the genotypic presence of co-resistance genes. Aminoglycoside and fluoroquinolone/quinolone resistance genes were the most frequently reported, each appearing in 15 studies, with aac(6′)-Ib-cr identified as the predominant resistance gene. Folate pathway antagonists’ resistance genes were reported in 12 studies, where dfrA14 was the most common resistance gene. This was followed by sulfonamides (11 studies), where sul2 was the most common gene, and tetracyclines (10 studies), with tet(A) as the predominant resistance gene. Phenicols were reported in 9 studies, with catA1 being the most prevalent gene. Finally, carbapenem resistance genes were documented in 8 studies, with blaNDM, blaOXA-48, and blaOXA-181 identified as the predominant resistance genes. Additionally, five studies reported mutations within the quinolone resistance-determining regions (QRDR) of gyrA, parC, parE, and parcE (Table 2). (Other reported co-resistance genes are present in the supplementary data 2; Table 16).
Mobile genetic elements
A total of 22 studies performed mobilome analyses to identify the plasmid types carrying ESBL and other co-resistance genes. IncF plasmids emerged as the most prevalent, found across human, animal, and environmental sources, with an overall prevalence of 50.0%. Col plasmids were the second most common, accounting for 16.4%. Other plasmid types, including IncH, IncQ, and IncI, were detected at lower frequencies, with prevalence rates ranging from ~ 4.0% to ~ 7.0%. (Fig. 7) (Supplementary data 2; Table 17–17A).
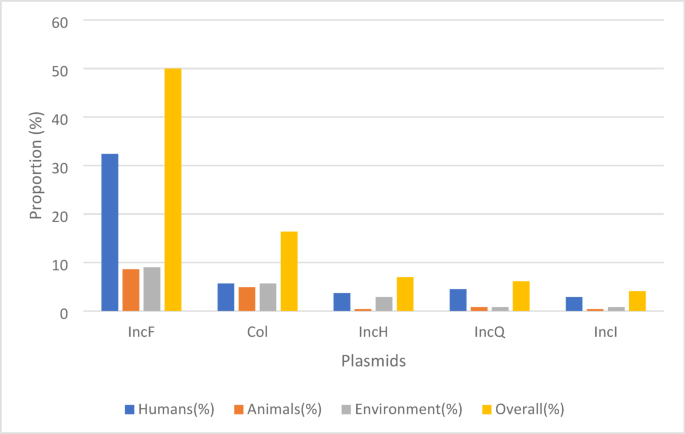
Prevalence of ESBL conjugative plasmids across human, animal, and environmental sources.
Meta-analysis
Prevalence of ESBL
The overall pooled prevalence of ESBL in West Africa was 16.8% (95% CI [12.1; 22.1]. Nigeria and Ghana reported the highest prevalence at 18.3% and 17.3%, respectively, while Burkina Faso and Senegal had low prevalences at 6.0% and 1.2%, respectively (Fig. 8).
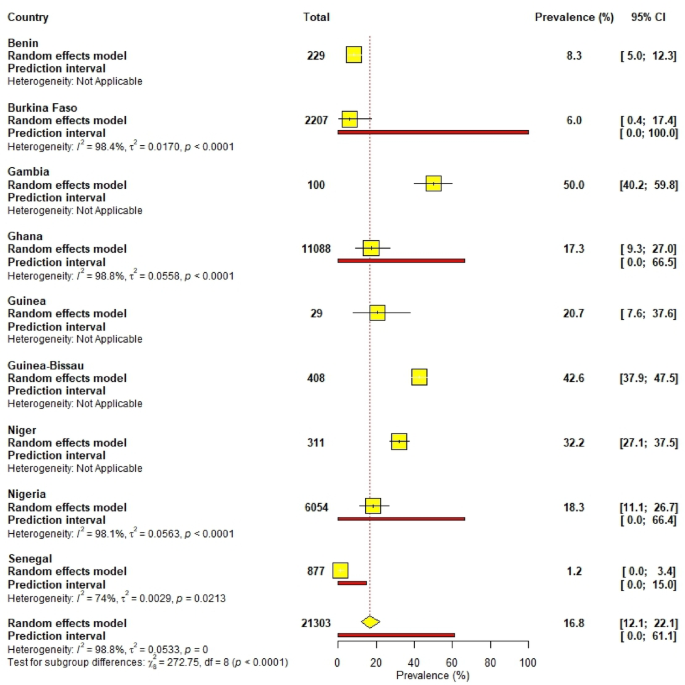
Pooled prevalence of ESBL-producing bacteria according to countries in West Africa.
Humans had the highest prevalence at 20.4%, followed by animals at 12.8% and the environment at 9.3% (Fig. 9). Among settings, market-based environments recorded the highest prevalence at 23.6%, followed by hospitals at 21.8% and community-based settings at 7.5% (Supplementary data 1; Fig. 1).
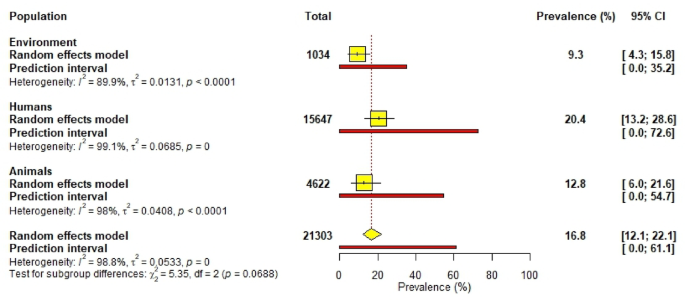
Pooled prevalence of ESBL-producing bacteria across human, animal, and environmental sources.
Prevalence of ESBL-producing E. coli and K. pneumoniae
The study explored ESBL-producing E. coli and K. pneumoniae in 34 of 38 and 20 of 38 studies, respectively, across humans, animals, and the environment (Supplementary data 2; Tables 18-23). The prevalence of ESBL E. coli was 15.9% (95% CI: [9.8; 23.1], 26 studies) in humans, 15.0% (95% CI: [6.5; 26.0], 11 studies) in animals, and 6.1% (95% CI: [1.8; 12.4], 5 studies) in the environment (Table 3). The prevalence of ESBL K. pneumoniae was 8.2% (95% CI: [3.9; 13.8], 16 studies) in humans, 2.0% (95% CI: [0.4; 4.3], 4 studies) in animals, and 6.6% (95% CI: [4.4; 9.2], 2 studies) in the environment (Table 4).
Subgroup analysis
The subgroup analysis of pooled prevalence estimates for ESBL-positive E. coli and K. pneumoniae across humans, animals, and the environment revealed distinct patterns based on settings and sample types.
For E. coli, human studies showed the highest prevalence in hospital-based settings (17.6%) and faecal samples (23.2%). Among animals, faecal samples have the highest prevalence (27.0%), and farm-based settings (21.7%) were particularly affected. In environmental studies, the abattoir environment has the highest rate (22.4%), whereas hospital-based environments have much lower rates (1.7%) (Table 3).
Clinical specimens: urine, ear swab, sputum, blood culture, eye swab, wound swab, urethral swab, endocervical swab, stool, high vaginal swab, nasal swab, gastric lavage, pus, tracheal aspirates, semen, axilla, groin, perianal region, cerebrospinal fluid, lower respiratory tracts, urinary tract infections (UTIs), ascitic fluid.
Patient environment: swabs of high- and low-touch areas, beds, taps, and drip stand.
NICU environment: swabs of incubator doors, cots, nurse trolley handle, weighing scales, tables, and desks.
For K. pneumoniae, human studies revealed a dominance of hospital-based settings (10.4%) and clinical specimens (9.5%) prevalence, with wound swabs reporting the highest prevalence in individual samples (57.1%). In animals (4 studies, 2.0%), community-based settings (2.5%), and rectal swabs (10.3%) showed higher prevalence rates. Environmental studies for K. pneumoniae (6.6% prevalence) indicated that patient environments (7.4%) had a slightly higher prevalence than NICU environments (5.8%) (Table 4).
Heterogeneity was significant in human studies on both isolates (I² ~99%), indicating variability across settings and sample types, whereas environmental studies on K. pneumoniae showed minimal variability (I² = 0%). This subgroup analysis indicated significant differences in the prevalence of ESBL E. coli and K. pneumoniae depending on the host/source, settings, and sample type.
Heterogeneity and publication bias
The funnel plot illustrates a publication bias, which was confirmed by Egger’s regression test (p = < 0.0001) (Supplementary data 1; Fig. 2). Estimated prevalences were significantly heterogeneous across countries, populations, and settings (H > 1 and I2 ≥ 75%). The overall prevalence also showed significant heterogeneity (H = 8.98 [8.51; 9.49] and I = 98.8% [98.6%; 98.9%]).
Sensitivity analysis and meta-regression
The impact of each study on the pooled prevalence of ESBL-producing bacteria was assessed using a leave-one-out sensitivity analysis. The sensitivity analysis revealed that excluding individual studies had a minimal impact on the pooled prevalence of 16.8%, with most exclusions resulting in prevalence values ranging from 16.0 to 17.4%. Only one study reported a slightly lower prevalence of 15.8%. These findings indicate that the pooled prevalence was stable and not overly influenced by any single study, demonstrating the robustness of the meta-analysis results (Supplementary data 1; Table 5).
A random effects meta-regression analysis was used to assess the effect of the three study characteristics on the observed wide variations in effect sizes in this study. Country, population, and settings were used as covariates. Gambia had a minimally significant negative association with heterogeneity. The remaining covariates did not significantly affect heterogeneity (Table 5).