Introduction: This study reports a confirmed case of human rabies diagnosed through metagenomic next-generation sequencing (mNGS). The patient was a 22-year-old female who developed symptoms 3 months after sustaining a scratch on the upper lip from a domesticated dog, without receiving postexposure prophylaxis (PEP). She initially presented with urinary symptoms and was misdiagnosed with a urinary tract infection. Neurological symptoms subsequently emerged, prompting intensive life support interventions including mechanical ventilation, recombinant human interferon-α2b, ribavirin, norepinephrine, veno-arterial extracorporeal membrane oxygenation (VA-ECMO), and continuous renal replacement therapy.
Methods: Clinical data were collected from hospital records, including exposure history, symptoms, treatments, and outcomes. Saliva specimens were tested by reverse transcription polymerase chain reaction (RT-PCR) and mNGS. Sequencing data were processed by standard bioinformatics pipelines, and phylogenetic analysis was performed with MAFFT alignment and IQ-TREE maximum-likelihood reconstruction.
Results: Rabies virus infection was confirmed through reverse transcription polymerase chain reaction (RT-PCR) and mNGS analysis of saliva samples. The detected strain, JSTZ190314, represents the first documented case of this genotype in Guangxi, China. Despite initial stabilization with ECMO support, the patient’s neurological condition deteriorated progressively, leading to brain death 28 days after neurological onset (34 days from initial urinary symptoms).
Conclusion: mNGS proves invaluable as a diagnostic tool for atypical rabies presentations. Enhancing early clinical recognition capabilities, ensuring timely and standardized PEP implementation, and strengthening regional viral strain surveillance represent critical components for effective rabies prevention and control strategies.
Rabies represents a fatal zoonotic disease caused by the rabies virus (RABV), resulting in over 59,000 human deaths globally each year, with the majority of cases occurring in Asia and Africa (1–2). Early diagnosis of rabies remains challenging due to the low detection rates of viral RNA in saliva and cerebrospinal fluid (CSF) during the initial stages of infection (3). Although the direct fluorescent antibody (DFA) test continues to serve as the criterion standard for rabies diagnosis, its sensitivity depends heavily on specimen quality, operator experience, and laboratory conditions. This variable sensitivity renders the DFA test susceptible to false-negative results, particularly in patients with low viral loads or early-stage infections. Similarly, polymerase chain reaction (PCR) methods have enhanced detection capabilities but are not without limitations, particularly in resource-constrained settings where accessibility and reproducibility may be compromised (4–5).
More recently, metagenomic next-generation sequencing has emerged as a powerful, unbiased, high-throughput tool for the rapid identification of infectious pathogens, particularly in cases involving rare, atypical, or difficult-to-detect organisms (6–7). Here, we report a patient with a rabies infection who survived 28 days from neurological onset (34 days from initial urinary symptoms) caused by the JSTZ190314 strain — the first such case in Guangxi Zhuang Autonomous Region, China — as confirmed via metagenomic next-generation sequencing. This report aims to highlight the diagnostic value of metagenomic next-generation sequencing in early rabies detection, describe the clinical course under aggressive supportive care, and explore the public health implications of a newly identified RABV strain.
Clinical data were obtained from hospital medical records, including demographics, exposure history, clinical manifestations, laboratory findings, imaging, and therapeutic interventions.
Saliva samples were tested for rabies virus RNA using reverse transcription polymerase chain reaction (RT-PCR). Metagenomic next-generation sequencing (mNGS) was performed on the GeneMind FASTASeq 300 platform (single-end 75 bp, >20 million reads per sample, Q30 ≥85%). Data were processed with fastp for quality control, low-complexity reads were filtered with PRINSEQ, and host sequences were removed by alignment to the hg38 human reference genome using BWA-MEM. Non-host reads were taxonomically classified with Kraken2 and validated by re-alignment.
Complete N gene sequences were aligned using MAFFT v7.490 (–auto), and phylogenetic reconstruction was conducted with IQ-TREE v2.0.7 (GTR+G model, 1,000 ultrafast bootstrap replicates). Visualization was performed with iTOL v7.2.1.
A previously healthy 22-year-old woman developed symptoms 3 months after sustaining a scratch on her upper lip from a domesticated dog in September 2024. Although the wound bled significantly, she only rinsed it with tap water and received neither rabies vaccination nor passive immunization.
On December 22, 2024, she experienced acute urinary frequency, urgency, and vulvar burning. Three days later, she visited a local hospital, where clinicians initially diagnosed urinary tract infection and vulvitis. Despite empirical antimicrobial therapy and symptomatic treatment, her condition showed no improvement. On December 28, the patient developed fever (38.2 °C), chills, pharyngeal spasms, muscle twitching, dysphagia, and pronounced anxiety. She presented to our emergency department where rabies was suspected, prompting immediate admission to the intensive care unit due to hemodynamic instability.
Upon admission, the patient exhibited agitation, hypersalivation, and an exaggerated pharyngeal reflex. She demonstrated marked hydrophobia and aerophobia with extreme sensitivity to auditory and airflow stimuli. Her vital signs revealed instability: heart rate 145 beats/min, blood pressure 90/50 mmHg, and peripheral oxygen saturation (SpO2) of 88% on room air. Laboratory findings showed leukocytosis (15.8×109/L) and elevated myocardial enzymes [creatine kinase (CK)-MB: 250.1 U/L; troponin T: 0.487 ng/mL]. Echocardiography revealed globally reduced cardiac systolic function with a left ventricular ejection fraction (LVEF) of 16%, suggesting concurrent viral myocarditis.
Due to rapid neurologic deterioration and circulatory collapse, the patient underwent emergency endotracheal intubation and mechanical ventilation on December 28. The following day, she progressed to cardiogenic shock despite high-dose norepinephrine infusion [3.5 μg/(kg·min)]. We urgently initiated veno-arterial extracorporeal membrane oxygenation (VA-ECMO) and continuous renal replacement therapy (CRRT) to stabilize vital signs. Concurrently, we administered antiviral therapy with ribavirin and recombinant interferon-α2b, along with corticosteroids, intravenous immunoglobulin, and comprehensive supportive care.
Following standard laboratory protocols for rabies diagnosis, we initially planned to collect multiple saliva specimens for testing. However, the first specimen collected on January 3, 2025, already tested positive for rabies virus RNA by RT-PCR (Ct=26.38). Given the concordant clinical presentation, we submitted this same sample for metagenomic next-generation sequencing (mNGS) and did not pursue additional specimen collection. On January 5, mNGS confirmed the presence of RABV infection. We performed sequencing on the GeneMind FASTASeq 300 platform using single-end 75 bp reads, generating over 20 million raw reads per sample with Q30 scores ≥85%. After quality control with fastp and removal of low-complexity sequences, we aligned clean reads to the human reference genome (hg38) using BWA-MEM and excluded host reads. We then taxonomically classified non-host reads with Kraken2 and validated results by re-alignment to reference genomes. To filter potential contaminants, we used negative controls and a laboratory background database, considering only microbes with reads per million (RPM) ≥3× negative template control (NTC) as positive. We annotated antimicrobial resistance and virulence genes by mapping against the CARD and VFDB databases. Sequence alignment identified the strain as JSTZ190314, marking the first reported case of this genotype in Guangxi, China. The evolutionary relationship of the patient-derived strain is shown in Figure 1.
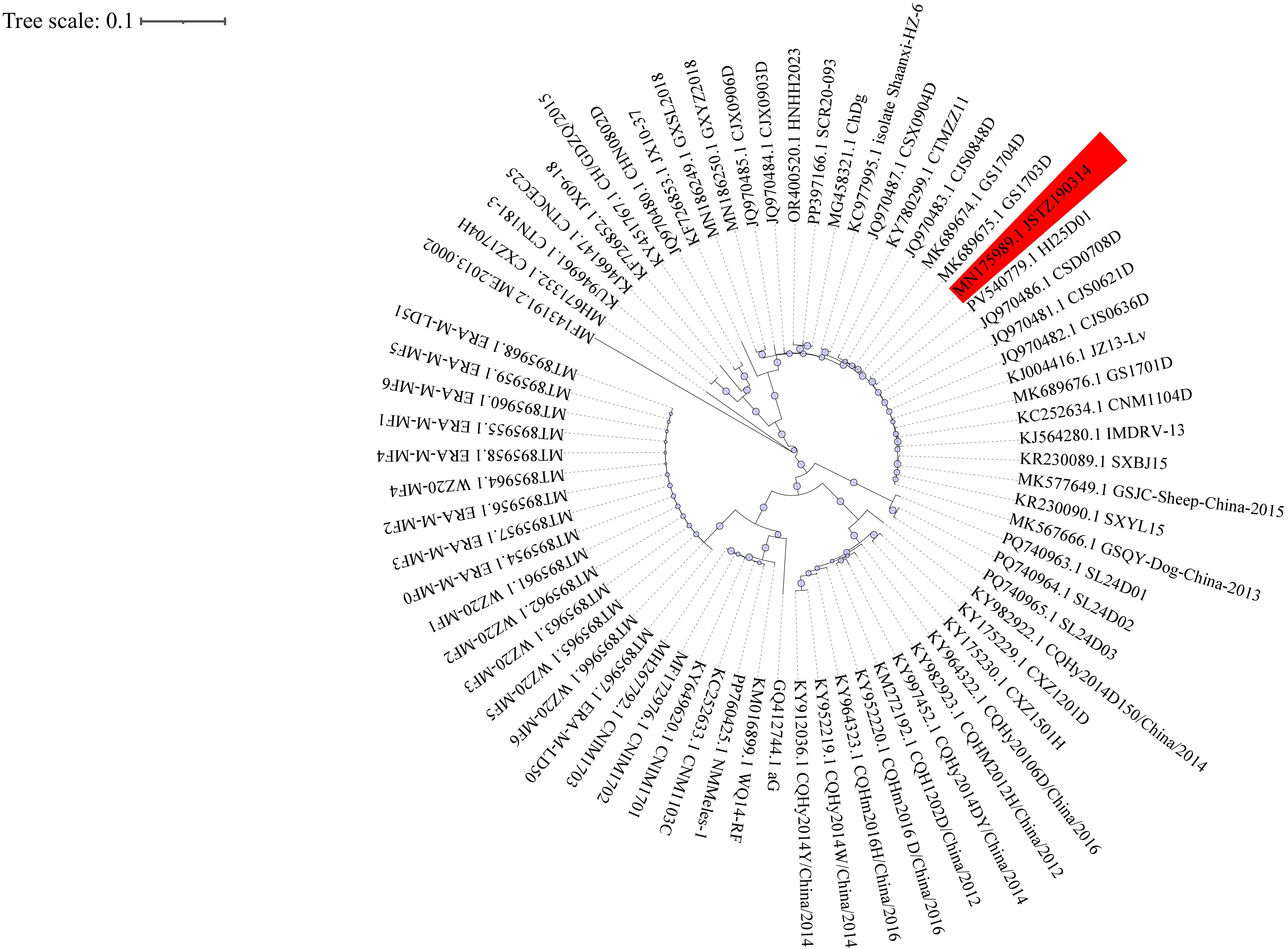
Phylogenetic analysis of Rabies lyssavirus strains in China.
Note: Circular maximum-likelihood phylogenetic tree of 70 complete N gene sequences of Rabies lyssavirus strains circulating in China. The tree was reconstructed using IQ-TREE v2.0.7with themaximum-likelihood methodunder theGTR+G substitution modeland1000 bootstrap replicates, based on multiple-sequence alignment performed by MAFFT v7.490 (–auto option). Bootstrap values (≥70) are shown at major nodes. The patient-derived strain JSTZ190314 is highlighted, with host species and geographic origins annotated from GenBank records. Visualization was performed with iTOL v7.2.1. Abbreviation: MAFFT=Multiple Alignment using Fast Fourier Transform; IQ-TREE=software for phylogenetic inference using maximum likelihood.
The patient was successfully weaned off ECMO on January 7, but her neurological condition progressively deteriorated. She remained in a deep coma with no spontaneous respiration, bilaterally dilated and fixed pupils, and absent pupillary light reflexes. Neurological evaluation confirmed brain death. Following written informed consent from the patient’s legal guardian, we withdrew life-sustaining treatment on January 25, and the patient was declared clinically dead. The patient progressed to brain death 28 days after neurological symptom onset (34 days from initial urinary symptoms). Figure 2 summarizes the clinical course.
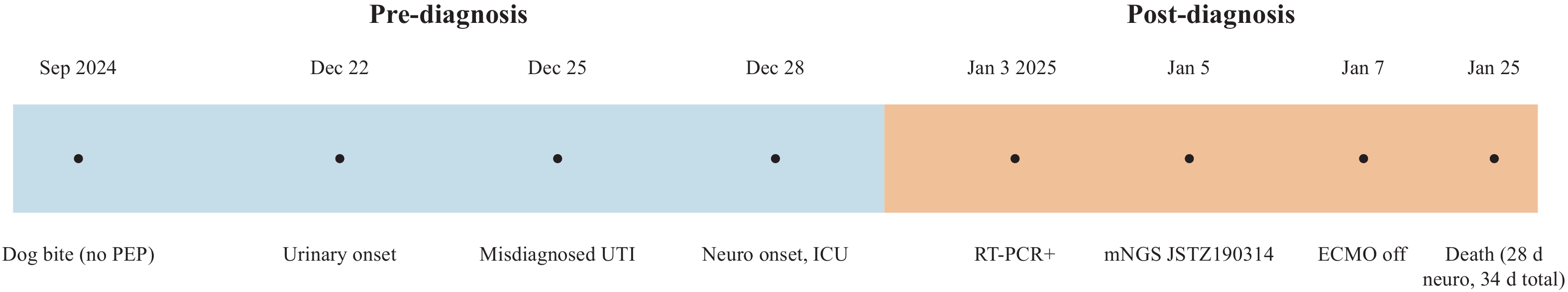
Clinical timeline of the reported rabies case.
Note: Pre-diagnosis events (blue): dog bite without PEP, urinary onset, misdiagnosis as UTI, and neurological onset with ICU admission. Post-diagnosis events (orange): saliva RT-PCR positive, mNGS confirmation of strain JSTZ190314, ECMO weaning, and death 28 days after neurological onset (34 days from initial symptoms).
Abbreviation: PEP=postexposure prophylaxis; UTI=urinary tract infection; mNGS=metagenomic next-generation sequencing; ECMO=extracorporeal membrane oxygenation.