European Commission. The European green deal. Publication Office of the European Union; 2019.
European Commission. Farm to fork strategy. 2020.
Glare T, Caradus J, Gelernter W, Jackson T, Keyhani N, Köhl J, et al. Have biopesticides come of age? Trends Biotechnol. 2012;30(5):250–8.
Google Scholar
Taning CNT, Mezzetti B, Kleter G, Smagghe G, Baraldi E. Does RNAi-based technology fit within EU sustainability goals?? Trends Biotechnol. 2021;39(7):644–7.
Google Scholar
Nunes PSO, Lacerda-Junior GV, Mascarin GM, Guimarães RA, Medeiros FHV, Arthurs S, et al. Microbial consortia of biological products: do they have a future? Biol Control. 2024;188:105439.
Wend K, Zorrilla L, Freimoser FM, Gallet A. Microbial pesticides – challenges and future perspectives for testing and safety assessment with respect to human health. Environ Health. 2024;23(1):49.
Google Scholar
European Commission. Guidance document on semiochemical active substances and plant protection products. 2024.
Scholz S, Brack W, Escher BI, Hackermüller J, Liess M, von Bergen M, et al. The EU chemicals strategy for sustainability: an opportunity to develop new approaches for hazard and risk assessment. Arch Toxicol. 2022;96(8):2381–6.
Google Scholar
Schmeisser S, Miccoli A, von Bergen M, Berggren E, Braeuning A, Busch W, et al. New approach methodologies in human regulatory toxicology – not if, but how and when! Environ Int. 2023;178:108082.
Google Scholar
Anisimov Andrey P, Lindler Luther E, Pier Gerald B. Intraspecific diversity of Yersinia pestis. Clin Microbiol Rev. 2004;17(3):695.
Belsham GJ, Kristensen T, Jackson T. Foot-and-mouth disease virus: prospects for using knowledge of virus biology to improve control of this continuing global threat. Virus Res. 2020;281:197909.
Google Scholar
Nitsche BM, Jørgensen TR, Akeroyd M, Meyer V, Ram AFJ. The carbon starvation response of Aspergillus Niger during submerged cultivation: insights from the transcriptome and secretome. BMC Genomics. 2012;13(1):380.
Google Scholar
Kusari S, Hertweck C, Spiteller M. Chemical ecology of endophytic fungi: origins of secondary metabolites. Chem Biol. 2012;19(7):792–8.
Google Scholar
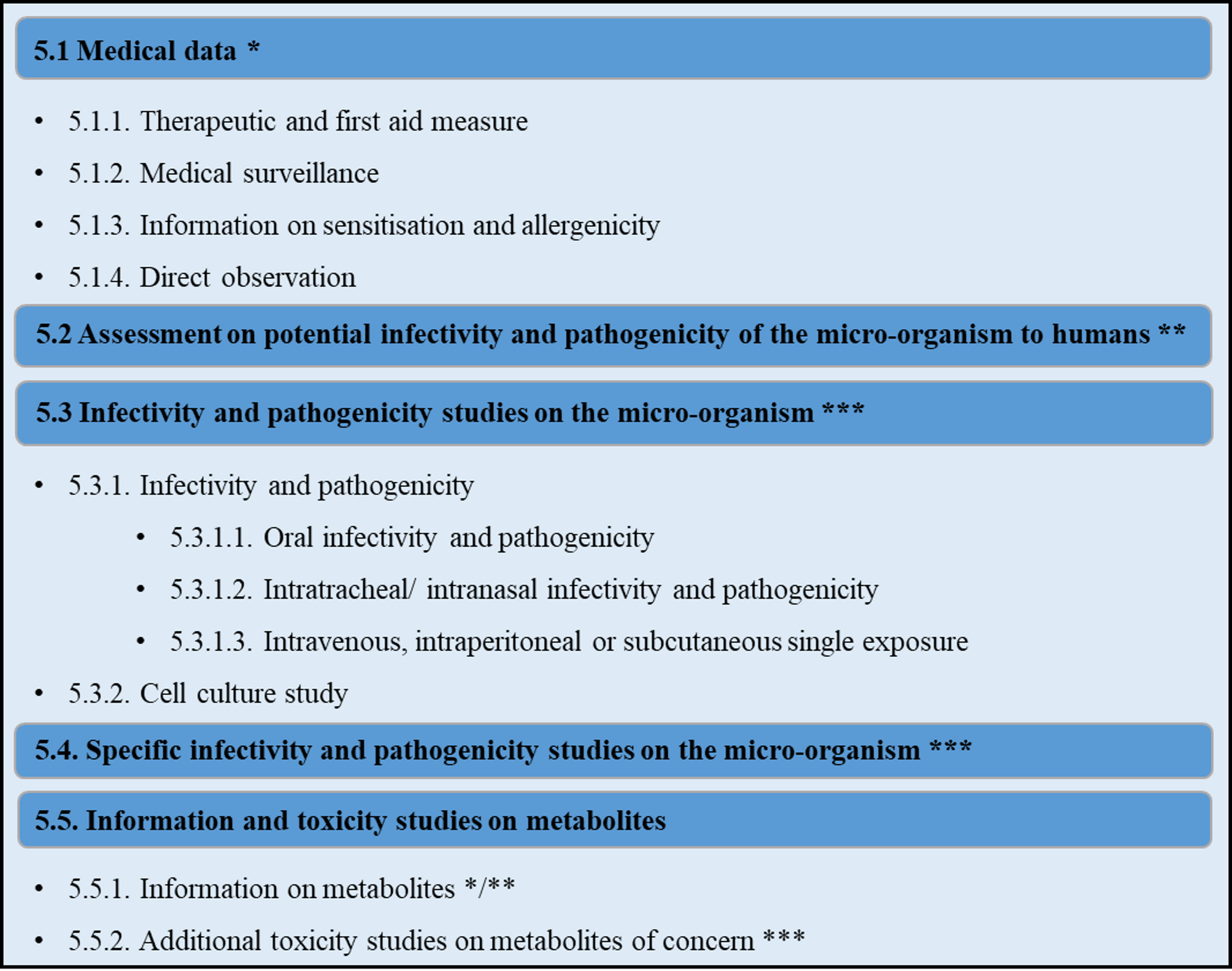
U.S. Environmental Protection Agency. Test guidelines for pesticides and toxic substances. Series 885 – Microbial pesticide test guidelines. Washington, DC: U.S. Environmental Protection Agency; 1996. Report No.: EPA Report No 885-XXX.
Tollefsen KE, Scholz S, Cronin MT, Edwards SW, de Knecht J, Crofton K, et al. Applying adverse outcome pathways (AOPs) to support integrated approaches to testing and assessment (IATA). Regul Toxicol Pharmacol. 2014;70(3):629–40.
Nymark P, Clerbaux L-A, Amorim M-J, Andronis C, de Bernardi F, Bezemer GFG et al. Building an adverse outcome pathway network for COVID-19. Front Syst Biology. 2024;4–2024.
Mayasich SA, Amorim M-J, Paini A, Parissis N, Kim HJ, Nymark P. Binding of SAR-CoV-2 to ACE2 leads to viral infection proliferation: AOP-Wiki/OECD; 2023 [Available from: https://aopwiki.org/aops/430.]
Busschers M, Gwynn R, Ramaekers L, Lewis J, Greco F. Data decision tree for identifying potential risks for natural substances when used in plant protection. Biocontrol Sci Technol. 2023;33(7):597–629.
Paege N, Feustel S, Marx-Stoelting P. Toxicological evaluation of microbial secondary metabolites in the context of European active substance approval for plant protection products. Environ Health. 2024;23(1):52.
Morais Leme D, Bomfim Pestana C, Kenny EF, Feustel S, Corsini E. Assessing the sensitisation hazard of microbial pesticides: potential value of new approach methodologies (NAMs) to overcome current challenges. Manuscript in preparation, 2025.
European Comission. Commission regulation (EC) no 1107/2009 concerning the placing of plant protection products on the market. Brussels: European Union; 2009.
European Commission. Commission implementing regulation (EU) 2022/1439 amending regulation (EU) 283/2013 as regards the information to be submitted for active substances and the specific data requirements for micro-organisms. Report No.: EU 2022/1439. Official J Eur Union. L 227/8, 2022.
European Commission. Commission implementing regulation (EU) 2022/1440 amending regulation (EU) no 284/2013 as regards the information to be submitted for plant protection products and the specific data requirements for plant protection products containing micro-organisms. Brussels: European Commission; 2022. Report No.: EU 2022/1440.
European Commission. Commission implementing regulation 2022/1441 amending regulation (EU) no 546/2011 as regards specific uniform principles for evaluation and authorisation of plant protection products containing micro-organisms. Brussels: European Union; 2022.
European Commission. Commission regulation (EU) no 547/2011 implementing regulation (EC) no 1107/2009 of the European Parliament and of the Council as regards labelling requirements for plant protection products. Brussels: European Union; 2011. Report No.: (EU) No 547/2011.
Panel Rychen EFSAFEEDAP, Aquilina G, Azimonti G, Bampidis G. Guidance on the characterisation of microorganisms used as feed additives or as production organisms. EFSA J. 2018;16(3):e05206.
EFSA. Submission of scientific peer-reviewed open literature for the approval of pesticide active substances under regulation (EC) 1107/2009. EFSA J. 2011;9(2):2092.
OECD. Innovating microbial pesticide testing: conference proceedings series on pesticides No. 109. Paris, France OECD. 2023. Report No.: ENV/CBC/MONO(2023)10.
Marx-Stoelting P, Rivière G, Luijten M, Aiello-Holden K, Bandow N, Baken K, et al. A walk in the PARC: developing and implementing 21st century chemical risk assessment in Europe. Arch Toxicol. 2023;97(3):893–908.
Google Scholar
Jacobs MN, Hoffmann S, Hollnagel HM, Kern P, Kolle SN, Natsch A, et al. Avoiding a reproducibility crisis in regulatory toxicology—on the fundamental role of ring trials. Arch Toxicol. 2024;98(7):2047–63.
Google Scholar
EFSA. EFSA statement on the requirements for whole genome sequence analysis of microorganisms intentionally used in the food chain. EFSA J. 2021;19(7):e06506.
U.S. Environmental Protection Agency. OPPTS 885.3050 Acute oral toxicity/ pathogenicity U.S. Environmental Protection Agency. 1996. Report No.: OPPTS 885.3050.
U.S. Environmental Protection Agency. OPPTS 885.3150 Acute pulmonary toxicity/ pathogenicity U.S. Enviromental Protection Agency. 1996. Contract No.: OPPTS 885.3150.
U.S. Environmental Protection Agency. OPPTS 885.3200 microbial pesticide test methods. Acute injection toxicity/pathogenicity. U.S. Environmental Protection Agency; 1996. Contract No.: OPPTS 885.3200.
U.S. Environmental Protection Agency. OPPTS 885.3500 cell culture. U.S. Environmental Protection Agency; 1996. Contract No.: OPPTS 885.3500.
U.S. Environmental Protection Agency. OPPTS 885.3600 subchronic toxicity/pathogenicity. U.S. Environmental Protection Agency; 1996. Contract No.: OPPTS 885.3600.
U.S. Environmental Protection Agency. OPPTS 885.3650 reproductive/fertility effects. U.S. Environmental Protection Agency; 1996. Contract No.: OPPTS 885.3650.
Anastassiadou EFSA, Arena M, Auteri M, Brancato D, Bura A. Peer review of the pesticide risk assessment of the active substance Bacillus thuringiensis subsp. Kurstaki strain SA-11. EFSA J. 2020;18(10):e06261.
Alvarez EFSA, Arena F, Auteri M, Binaglia D, Castoldi M. Peer review of the pesticide risk assessment of the active substance Aspergillus flavus strain MUCL54911. EFSA J. 2022;20(3):e07202.
European Commission. Directive 2010/63/EU of the European Parliament and of the Council on the protection of animals used for scientific purposes European Union; 2010. Report No.: Directive 2010/63/EU.
EFSA FEEDAP Panel. Guidance on the assessment of the toxigenic potential of Bacillus species used in animal nutrition. EFSA J. 2014;12(5):3665.
Fichant A, Lanceleur R, Hachfi S, Brun-Barale A, Blier A-L, Firmesse O et al. New approach methods to assess the enteropathogenic potential of strains of the Bacillus cereus group, including Bacillus thuringiensis. Foods. 2024;13(8).
Walocha R, Kim M, Wong-Ng J, Gobaa S, Sauvonnet N. Organoids and organ-on-chip technology for investigating host-microorganism interactions. Microbes Infect. 2024;26(7):105319.
Google Scholar
de Oliveira LF, Filho DM, Marques BL, Maciel GF, Parreira RC, doCN J.R., et al. Organoids as a novel tool in modelling infectious diseases. Semin Cell Dev Biol. 2023;144:87–96.
Yokoi F, Deguchi S, Takayama K. Organ-on-a-chip models for elucidating the cellular biology of infectious diseases. Biochimica et biophysica acta (BBA) -. Mol Cell Res. 2023;1870(6):119504.
Google Scholar
European Commission. SANTE/2020/12260. Guidance on the approval and low-risk criteria linked to antimicrobial resistance applicable to microorganisms used for plant protection. Accordance with regulation (EC) no 1107/2009. Brussels: European Union; 2020. Report No.: SANTE/2020/12260.
WHO. GLASS whole-genome sequencing for surveillance of antimicrobial resistance. 2020.
Cassini A, Högberg LD, Plachouras D, Quattrocchi A, Hoxha A, Simonsen GS, et al. Attributable deaths and disability-adjusted life-years caused by infections with antibiotic-resistant bacteria in the EU and the European economic area in 2015: a population-level modelling analysis. Lancet Infect Dis. 2019;19(1):56–66.
European Commission. Commission Regulation (EU) No. 283/2013 setting out the data requirements for active substances, in accordance with Regulation (EC) No 1107/2009 of the European Parliament and of the Council concerning the placing of plant protection products on the market. Report No.: (EU) No. 283/2013. Official J Eur Union. L 93/1, 2013.
European Commission. Commission regulation (EU) no 546/2011 implementing regulation (EC) no 1107/2009 of the European Parliament and of the Council as regards uniform principles for evaluation and authorisation of plant protection products. Brussels: European Commission; 2011.
Dang T, Loll B, Müller S, Skobalj R, Ebeling J, Bulatov T, et al. Molecular basis of antibiotic self-resistance in a bee larvae pathogen. Nat Commun. 2022;13(1):2349.
Google Scholar
Hutchings MI, Truman AW, Wilkinson B. Antibiotics: past, present and future. Curr Opin Microbiol. 2019;51:72–80.
Google Scholar
Molloy EM, Hertweck C. Antimicrobial discovery inspired by ecological interactions. Curr Opin Microbiol. 2017;39:121–7.
European Commission. Commission regulation (EU) 2017/1432 amending regulation (EC) 1107/2009 of the European Parliament and the Council concerning the placing of plant protection products on the market as regards the criteria for the approval of low-risk active substances. Report No.: (EU) 2017/1432. Official J Eur Union. L 205/59, 2017.
European Commission. Commission regulation (EU) 2022/1438 amending Annex II to regulation (EC) no 1107/2009 of the European Parliament and of the Council as regards specific criteria for the approval of active substances that are micro-organisms. Brussels: European Commission; 2022. Report No.: (EU) 2022/1438.
Patel S. Drivers of bacterial genomes plasticity and roles they play in pathogen virulence, persistence and drug resistance. Infect Genet Evol. 2016;45:151–64.
Google Scholar
Fitzpatrick DA. Horizontal gene transfer in fungi. FEMS Microbiol Lett. 2012;329(1):1–8.
Google Scholar
Stanczak-Mrozek KI, Laing KG, Lindsay JA. Resistance gene transfer: induction of transducing phage by sub-inhibitory concentrations of antimicrobials is not correlated to induction of lytic phage. J Antimicrob Chemother. 2017;72(6):1624–31.
Google Scholar
Panel EFSABIOHAZ, Allende A, Alvarez-Ordóñez A, Bolton D, Bover-Cid S, Chemaly M, De Cesare A, et al. Statement on how to interpret the QPS qualification on ‘acquired antimicrobial resistance genes’. EFSA J. 2023;21(10):1–13.
Waddington C, Carey ME, Boinett CJ, Higginson E, Veeraraghavan B, Baker S. Exploiting genomics to mitigate the public health impact of antimicrobial resistance. Genome Med. 2022;14(1):15.
Goodwin S, McPherson JD, McCombie WR. Coming of age: ten years of next-generation sequencing technologies. Nat Rev Genet. 2016;17(6):333–51.
Google Scholar
Zhang J-Y, Roberts H, Flores DSC, Cutler AJ, Brown AC, Whalley JP, et al. Using de novo assembly to identify structural variation of eight complex immune system gene regions. PLoS Comput Biol. 2021;17(8):e1009254.
Google Scholar
Ellington MJ, Ekelund O, Aarestrup FM, Canton R, Doumith M, Giske C, et al. The role of whole genome sequencing in antimicrobial susceptibility testing of bacteria: report from the EUCAST subcommittee. Clin Microbiol Infect. 2017;23(1):2–22.
Google Scholar
McArthur AG, Waglechner N, Nizam F, Yan A, Azad MA, Baylay AJ, et al. The comprehensive antibiotic resistance database. Antimicrob Agents Chemother. 2013;57(7):3348–57.
Google Scholar
Jia B, Raphenya AR, Alcock B, Waglechner N, Guo P, Tsang KK, et al. CARD 2017: expansion and model-centric curation of the comprehensive antibiotic resistance database. Nucleic Acids Res. 2017;45(D1):D566–73.
Google Scholar
Alcock BP, Raphenya AR, Lau TTY, Tsang KK, Bouchard M, Edalatmand A, et al. CARD 2020: antibiotic resistome surveillance with the comprehensive antibiotic resistance database. Nucleic Acids Res. 2020;48(D1):D517–25.
Google Scholar
Zankari E, Hasman H, Cosentino S, Vestergaard M, Rasmussen S, Lund O, et al. Identification of acquired antimicrobial resistance genes. J Antimicrob Chemother. 2012;67(11):2640–4.
Google Scholar
Gupta SK, Padmanabhan BR, Diene SM, Lopez-Rojas R, Kempf M, Landraud L, et al. ARG-ANNOT, a new bioinformatic tool to discover antibiotic resistance genes in bacterial genomes. Antimicrob Agents Chemother. 2014;58(1):212–20.