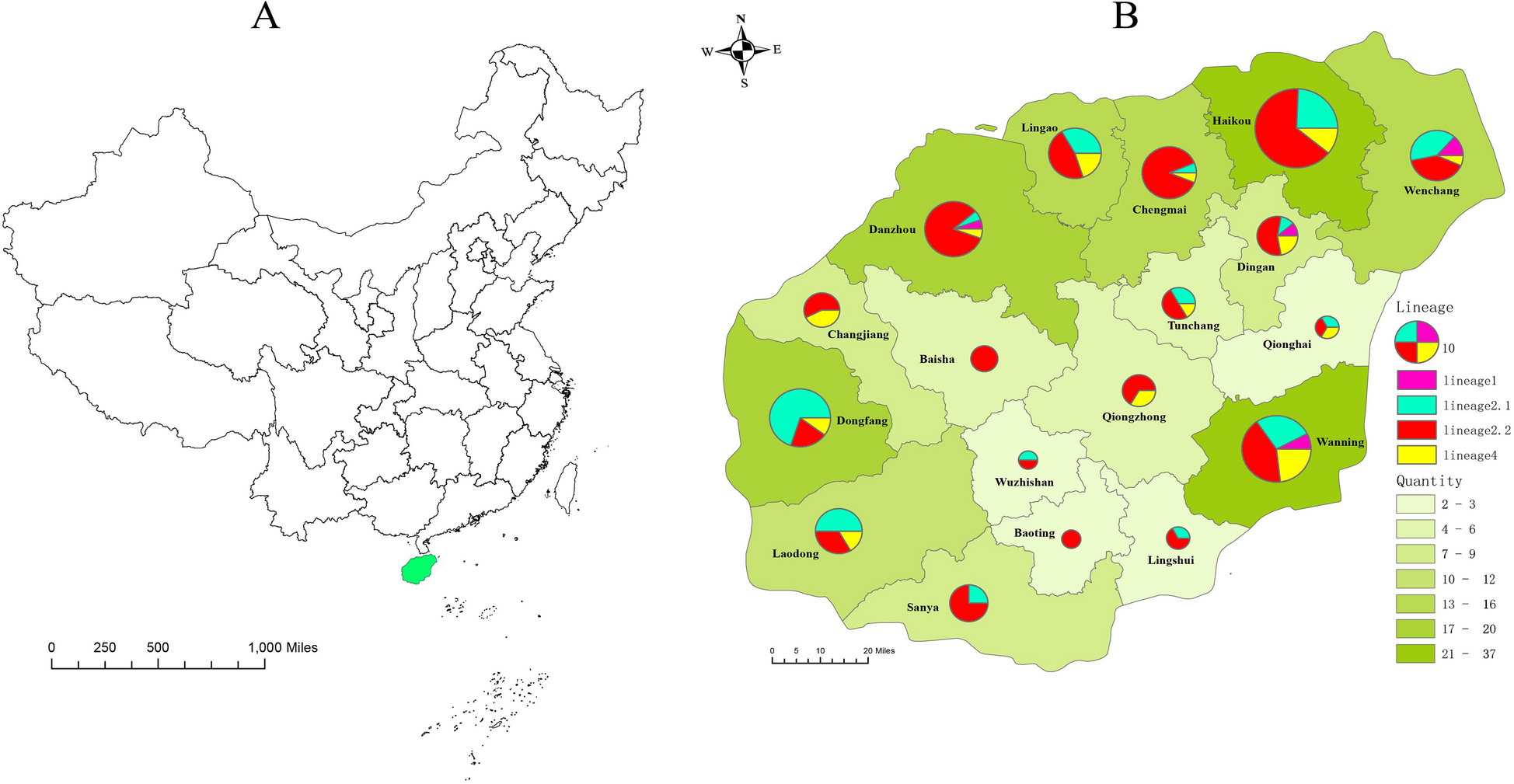
Bacterial strain data description
A total of 209 MDR-MTB strains were included in this study, originating from 18 cities and counties across Hainan Island, China. Haikou contributed the highest number of isolates (Fig. 1). The lineage distribution was as follows: 6 strains (2.9%) belonged to lineage 1, 57 strains (27.3%) to lineage 2.1, 117 strains (56.0%) to lineage 2.2, and 29 strains (13.9%) to lineage 4.
Distribution map of MDR-TB on the Hainan Island, China. A The geographical location of Hainan island in China. Green means Hainan Island. B Number and lineage distribution of MDR-TB patients on the Hainan Island
Correlation between lineage and phenotypic drug sensitivity
The MIC of all 209 MDR-MTB strains against 15 anti-TB drugs were determined using BMD plates. The observed resistance rates were as follows: 100.0% (209/209) for isoniazid (INH) and rifampicin (RIF), 41.1% (86/209) for ethambutol (EMB), 37.3% (78/209) for pyrazinamide (PZA), 42.6% (89/209) for moxifloxacin (MOX), 11.0% (23/209) each for amikacin (AMK) and kanamycin (KM), 9.6% (20/209) for capreomycin (CPM), 8.1% (17/209) each for protionamide (PTO) and para-aminosalicylic acid (PAS), 2.4% (5/209) for cycloserine (CS), 1.0% (2/209) for linezolid (LZD), and 0.5% (1/209) for delamanid (DLM). Although resistance to clofazimine (CFZ) and bedaquiline (BDQ) was not detected, their MIC exceeded the epidemiological cutoff values (ECOFFs).
For new or repurposed drugs like LZD, CFZ, BDQ, and DLM, the MIC50 and MIC90 values are ≤ 0.25 μg/mL and 0.5 μg/mL, ≤ 0.06 μg/mL and 0.5 μg/mL, ≤ 0.03 μg/mL and 0.06 μg/mL, ≤ 0.007 μg/mL and 0.03 μg/mL respectively. Lineage-based analysis revealed that MDR-MTB strains from lineage 2.2 exhibited a significantly higher resistance rate to EMB compared with non-lineage 2 strains (P < 0.05). Additionally, the mutation rates associated with PZA resistance in strains from lineages 2.1 and 2.2 were significantly higher than those in non-lineage 2 strains (P < 0.05) (Table 1, Fig. 2).
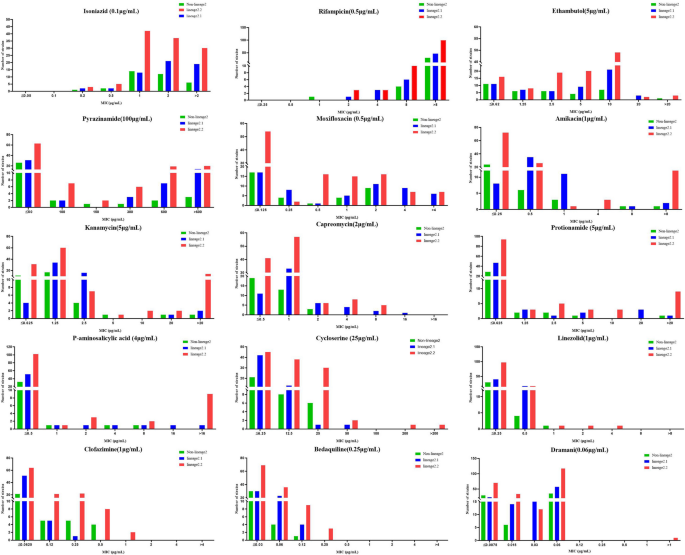
MIC distribution of 209 MDR-MTB strains
Correlation between Lineage and drug resistance gene mutation
By constructing a maximum likelihood phylogenetic tree linking strain lineages with anti-tuberculosis drug resistance (Fig. 3), we observed that lineage 2.2 strains exhibited significantly higher resistance rates to EMB compared with non-lineage 2 strains (P < 0.05). Lineage 2.2 strains also showed a higher resistance rate to pyrazinamide (PZA) compared with lineage 2.1 (P < 0.05).
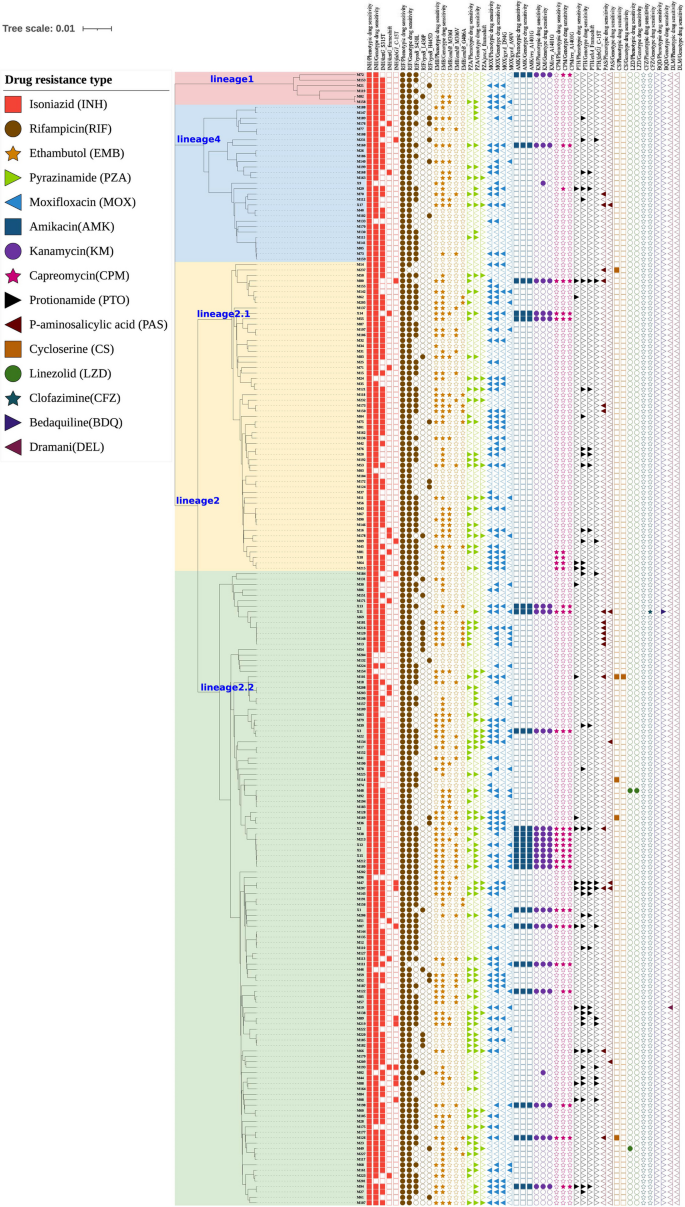
Maximum likelihood phylogenetic Tree of 209 MDR-MTB strains on the Hainan Island, China. Drug mutations are represented by patterns of different colors. Mutations are represented by filled (with mutations) or empty (without mutations) symbols
Regarding resistance-associated mutations, the embB_M306I mutation (for EMB) and gyrA_D94G mutation (for MOX) were more prevalent in non-lineage 2 and lineage 2.1 strains, respectively. In contrast, lineage 2.2 strains were characterized by the embB_M306V and gyrA_A94V mutations associated with resistance to EMB and MOX, respectively (Supplementary Table 3).
Comparison of consistency between BMD method and WGS drug sensitivity
For the first-line drugs (INH, RIF, EMB, and PZA) the sensitivity of WGS in predicting resistance was 94.7%, 99.0%, 96.5%, and 80.8%, respectively. However, the specificity for EMB and PZA was relatively low, at 60.2% and 79.4%, respectively. For second-line injectable aminoglycosides, including AMK, KM, and CPM, both sensitivity and specificity exceeded 95.0%. Apart from MOX, which showed a sensitivity of 94.4%, the sensitivity for other second-line drugs was below 80.0% (Table 2).
The relationship between different resistance-associated mutations and MIC
To evaluate the relationship between resistance-associated gene mutations and drug resistance levels, we analyzed 209 MDR-MTB strains from Hainan Island, China. MIC values for 15 anti-TB drugs were compared with corresponding resistance-associated mutations. The analysis revealed that specific mutations were associated with distinct MIC ranges (Supplementary Table 4).
For the first-line drug INH, non-synonymous mutations in katG were generally associated with high-level resistance (MIC ≥ 1 µg/mL). However, strains harboring katG_S315T (8 strains), Y337C (1 strain), N138D (1 strain), and P232R (1 strain) exhibited low-level resistance. Mutations in the fabG1 and ahpC promoter regions were also linked to low-level INH resistance, though co-occurrence of both mutations often correlated with increased MIC and high-level resistance.
For RIF, mutations were frequently observed at codons 435, 445, and 450 of rpoB. Double mutations in rpoB typically conferred high-level resistance (MIC ≥ 8 µg/mL), while some strains with single mutations such as D435F (2 strains), D435V (1 strain), D435Y (1 strain), H445S (1 strain), and L430P + H445Q showed low-level resistance (MIC ≤ 4 µg/mL). Two strains exhibited insertion-deletion mutations in rpoB (1295_1303_delAATTCATGG and R384W + 1299_1304_delCATGGA), and one strain with a V170F mutation located outside the RIF resistance-determining region showed high-level resistance.
EMB resistance-associated mutations were primarily located in embB and were linked to MIC near the ECOFF (5 µg/mL), indicating low-level resistance or phenotypic susceptibility. Strains with promoter mutations in embA typically had MIC ≤ ECOFF, suggesting susceptibility. However, when embA and embB mutations co-occurred, MIC tended to increase, indicating low-level resistance.
For PZA, resistance-associated mutations were distributed throughout the pncA gene and its promoter, including diverse single-nucleotide polymorphisms (SNPs) and insertions/deletions. These mutations were highly heterogeneous and lacked specific hotspots, consistent with previous reports [25, 26]. A frameshift mutation in the pncA gene led to a modest increase in mutation frequency. Additionally, different types of resistance-associated mutations were found to occur within various minimum inhibitory concentration (MIC) ranges. Interestingly, the MIC values for some wild-type strains were higher than the epidemiological cutoff values (ECOFF). Interestingly, some wild-type strains showed MIC exceeding the ECOFF, making it difficult to establish a clear correlation between pncA mutations and resistance levels.
Among second-line drugs, most fluoroquinolone-resistant strains harbored mutations in gyrA or gyrB. The most common gyrA mutations occurred at codons 90 and 94. gyrA_94 mutations were generally associated with high-level MOX resistance (MIC ≥ 2 µg/mL), while gyrA_90 mutations conferred low-level resistance or susceptibility. gyrB mutations were associated with MIC ≤ ECOFF, although combined gyrA/gyrB mutations often conferred high-level resistance.
Resistance to aminoglycosides was predominantly linked to the rrs_A1401G mutation, which typically conferred high-level resistance to AMK and KM, but only low-level resistance or even susceptibility to CPM, indicating that rrs_A1401G alone may not be predictive of CPM resistance.
The association between mutations in fabG1, the inhA promoter, and the ethA shift region with resistance to PTO was unclear. For PAS, mutations were primarily located in folC and thyA, but the link between these mutations and resistance was inconclusive, possibly due to unreliable phenotypic DST results.
Similarly, for CS, phenotypic DST was inconsistent, although one ald_433_ins_GC mutation was associated with high-level resistance. A single rplC_C154R mutation conferred high-level resistance to LZD. Additionally, an Rv0678_192_ins_G mutation led to cross-resistance to CFZ and BDQ, although phenotypic testing indicated MIC remained ≤ ECOFF for both drugs. No gene mutations associated with DLM resistance were detected.